
Complete Genome Sequence and Characterization of Beet Western Yellows Virus in Figwort
 ; Joong Hwan Lee2
; Joong Hwan Lee2 ; Yong Ho Jung3, #
; Yong Ho Jung3, # ; Da Hyun Lee4
; Da Hyun Lee4 ; Jun Hyeok Kim5
; Jun Hyeok Kim5 ; Chae Sun Na6
; Chae Sun Na6 ; Chung Youl Park7, †
; Chung Youl Park7, †
This is an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Figwort (Scrophularia buergeriana Miq.) is a biennial plant, whose dried root has been used in traditional medicine. Here, we investigated a viral disease in S. buergeriana, characterized the disease symptoms, and sequenced the complete genome of the identified virus.
Disease symptoms and growth characteristics were compared between two groups of plants, that is, those that were grown from rootlets and those cultivated from seeds. Leaf samples were collected from both groups for RNA sequencing analysis. Primers were designed based on the obtained contig, and the complete genome sequence was determined via reverse trancription polymerase chain reaction and cloning. Viral symptoms were confirmed in figwort plants. The highest incidence of infections was observed at the end of July. The complete genome of Beet western yellows virus (BWYV) ADHS comprised 5,878 nucleotides and seven open reading frames. Phylogenetic analysis based on the nucleotide sequence of the complete genome confirmed that the BWYV ADHS isolate was related most closely to the LS isolate from Korea.
This study is the first report of BWYV infection in figwort worldwide. The virus affects plant height and root weight. Control methods should be developed to minimize virus damage in figwort culture.
Keywords:
Scrophularia buergeriana, Beet Western Yellows Virus, Bionomical Characteristics, Polerovirus, RNA SequencingINTRODUCTION
Figwort (Scrophularia buergeriana Miq.) from the Scrophulariaceae family is a biennial to perennial herbaceous plant distributed in eastern Asia (Korea, Japan, and northern China) (Scheunert and Heubl, 2011).
The dried roots of figwort plants have been used in herbal medicine to treat various diseases (i.e., neuritis, sore throat, and laryngitis) (Kim et al., 2012), and they contain ingredients such as scrophularin, saikosaponins, iridoid glycosides, phenylpropanoids, terpenoids, and flavonoids (Li et al., 2000; Kim et al., 2009a). Of these, saikosaponins have been reported to have antiviral activity against HCoV-22E9, a coronavirus species infecting humans (Cheng et al., 2006).
Figwort propagation commonly uses seedlings or rootlets rather than seeds as the source material, as it can shorten the cultivation time by one year (Park et al., 2003).
Beet western yellows virus (BWYV) belongs to the genus Polerovirus in the family Luteoviridae. The genome of the virus is the positive-sense monopartite RNA 5.6 - 6.0 kb long with a genome-linked protein (VPg) at the 5' end (Peng et al., 2019; Candresse et al., 2020). The Polerovirus genome contains seven open reading frames (ORFs), including the recently identified small non-AUG-initiated ORF3a. ORF3a is located upstream of ORF3, and its protein is required for long-distance movement of the virus (Smirnova et al., 2015). This virus has icosahedral particles and is transmitted by aphids in a circulative, non-propagative manner (Knierim et al., 2014). BWYV has a broad host range and causes viral symptoms through phloem limitation (Reinbold et al., 2001).
In Korea, BWYV was first reported in paprika (Capsicum annuum var. angulosum) and later in hot pepper and various weed plants (Kwon et al., 2016; 2018; Park et al., 2018). BWYV was determined to have settled in the domestic ecology of Korea and was recently removed from the list of quarantine viruses. Nevertheless, this virus can infect monocotyledonous and dicotyledonous plants (Pagán and Holmes, 2010; Domier, 2012), causing economic damage, and requires continuous outbreak monitoring.
This study examined figwort plants exhibiting viral symptoms that were grown in a medicinal exhibition field at the Institute for Bioresources Research in Gyeongsangbuk-do, Korea. We identified BWYV for the first time from figwort plants, described the disease symptoms, compared growth characteristics of plants, and determined the complete genome sequence of the BWYV ADHS isolate.
MATERIALS AND METHODS
1. Plant material
To confirm the bionomical characteristics of figwort (Scrophularia buergeriana Miq.) plants, two groups (seed and root) were planted in an open field from 2017 to 2020 in Andong, Gyeongsanbuk-do Province. The seed group comprised figwort plants germinated from the seeds and grown in a plastic seed tray for 25 days, whereas the root group comprised plants grown from rootlets that were planted directly in the field.
Virus symptoms were investigated from mid-May to the end of July in 2017 and 2018. In addition, 20 virus-infected and symptomless plants were selected to measure and compare plant height, diameter, number of roots, and root weight. The data were analyzed using Duncan’s Multiple Range Test (DMRT), with significant difference considered at p < 0.05.
2. Total RNA extraction and RNA sequencing
To identify the causal agent(s) of the virus disease that produced mosaic symptoms in figwort plants, 50 leaf samples from the 200 samples leaf samples were collected from both groups in 2019 and 2020. All collected leaf samples were pooled and macerated with a mortar and pestle using liquid nitrogen.
Total RNA from the pooled sample was extracted using a Maxwell® RSC plant RNA kit (Promega Co., Madison, WI, USA) according to the manufacturer’s instructions and used for RNA sequencing (RNA-Seq) analysis. Ribosomal RNA depletion of total RNA was performed using a Ribo-Zero RNA removal kit, and the library was constructed using a TruSeq Stranded Total RNA LT Sample Prep Kit (Plant) (Illumina Inc., San Diego, CA, USA).
Sequencing was performed using the 2 × 101 bp paired-end method on an Illumina NovaSeq 6000 platform (Illumina Inc., San Diego, CA, USA). The obtained raw sequence data were processed and analyzed as previously described (Lee et al., 2020a). RNA-Seq and the analysis were performed by Macrogen (Macrogen, Seoul, Korea).
3. Validation of virus contig
To confirm the viral infection by BWYV, total RNA was extracted from all collected individual samples using an Easyspin total RNA extraction kit (iNtRON Bio Inc., Daejeon, Korea).
Two virus detection primer pairs (BWYV-F: 5'-GAA-ATT-GAA-TCA CCG-ACA-CGA-3' and BWYV-R: 5'-GCT-TGC-TTT-TCC-TTT ATG-AGC-3'; PaMMV-F: 5'-TTG-AGG-GTG-TTT-GCA-CTG-AAT-3' and PaMMV-R: 5'-TTG ACG-TGG-TAC-CTC-GTG-AA-3') were designed based on each contig sequence. One-step reverse transcription PCR (RT-PCR) was performed using a SuPrimeScript RT-PCR Premix (GenetBio, Daejeon, Korea). The amplicons were separated by 1% agarose gel electrophoresis, stained with EcoDye (Biofact, Daejeon, Korea), and observed using a UV illuminator (Major Science, Saratoga, CA, USA).
All fragments were directly sequenced in both directions using the ABI PRISM 3730XL analyzer with specific primer pairs, and the sequences were confirmed using NCBI BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
4. Genome and phylogenetic analysis
Of the samples with BWYV infection, one sample was randomly selected to determine the whole genome sequence. First-strand complementary DNA (cDNA) was synthesized from previously extracted total RNA with random N25 primers using MMLV reverse transcriptase (Invitrogen, Waltham, MA, USA) as outlined by the manufacturer.
To amplify the whole genome sequence of BWYV, six primers were designed based on contig sequence information (Table 1). PCR amplification was performed in a 20 ㎕ reaction mixtures containing 2 × TOPsimple DyeMIX (aliquot)-nTaq (Enzynomics Inc., Seoul, Korea) and 10 p㏖ of each primer set. The PCR reaction was performed in a Gene-Explorer thermal cycler (Bioer Co., Ltd., Hangzhou, China) using the following cycling parameters: 94℃ for 5 min; 32 cycles of 94℃ for 30 s, 55℃ for 30 s, and 72℃ for 1 min 30 s; and final extension at 72℃ for 5 min.
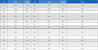
Primer information for complete genome sequence of Beet western yellows virus (BWYV) used in this study.
The 5' terminus was determined using terminal deoxynucleotidyl transferase (Takara, Tokyo, Japan) and gene-specific primers (GSP-R1: 5'-TTG-TAA-CTG-CTC-ACT-AAG-GAG-3' and GSP-R2: 5'-TCG-TAA-ATC-AAC-TGC-GCT-TGA-3') according to the 5' RACE System for Rapid Amplification of cDNA Ends, Version 2.0 (Roche, Mannheim, Germany). The 3' terminal sequence was generated using a poly(A) polymerase tailing kit (Lucigen Co., Middleton, WI, USA) with specific primers (GSP-F1: 5'-ATC-GAG-TGT-CCT-CAA-ACC-TC-3' and GSP-F2: 5'-CAG-CCA-CTA-TCA-TTG-CA-3') according to the manufacturer’s instructions. PCR amplification and gel electrophoresis analyses were carried out using the abovementioned methods.
All PCR products were cloned into T vector and sequenced as described previously (Park et al., 2016). The obtained sequences were edited and assembled using DNAMAN software package (Version 5.2.2, Lynnon Biosoft, San Ramon, CA, USA). The ORF was predicted using the NCBI ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/).
5. Identity comparison and phylogenetic analysis
Phylogenetic analysis based on complete genome sequences was performed to examine the relationships between the ADHS isolate and previously reported BWYV isolates. Complete genome sequences of 13 BWYV isolates and two species (Citrus vein enation virus and Soybean dwarf virus) as outgroups were retrieved from NCBI GenBank. Neughbor-joining phylogenetic tree and identity comparisons were generated using DNAMAN software.
RESULTS
1. Symptoms and bionomical characteristics
Viral infection symptoms in figwort (Scrophularia buergeriana Miq.) plants grown in the field were continuously detected in two consecutive years. The main symptoms of the predicted viral infection in plant samples included dwarfism, mosaic pattern, and yellowing of the leaves (Fig. 1).

Figwort (Scrophularia buergeriana) plants naturally infected with a virus. (A) Leaves showing dwarf, mosaic, and yellowing symptoms; (B) Enlarged section of A.
In both groups, the greatest number of symptoms was observed at the end of July (Fig. 2), and starting from August, the symptoms were masked, thus impeding their diagnosis with the naked eye.
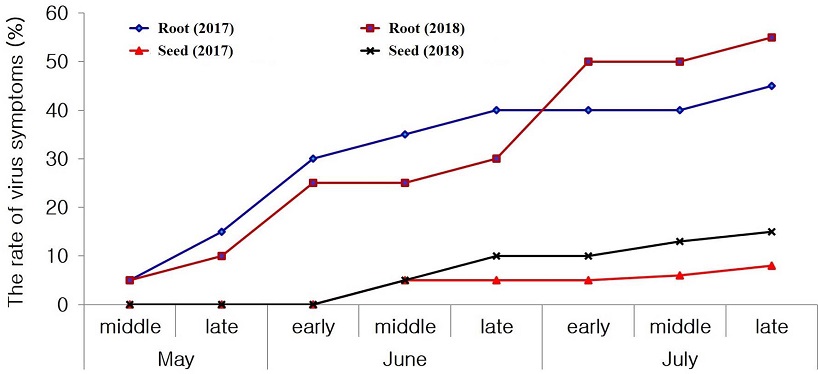
Incidence rate of virus-like symptoms on figwort plant leaves in the open filed during 2017 and 2018. Seed group; figwort seeds were sown and grown in a plastic tray and after 25 days transplanted into the open field. Root group; rootlets of figwort plants were directly planted in the open field. The field is located at the Institute for Bioresources Research in Gyeongsangbuk-do, Korea.
The greatest difference in growth parameters was observed for plant height and root weight. Healthy plants were taller and the roots were heavier compared to those of virus-infected samples, whereas the difference between the plant diameter and the root number between the two was not significant (Table 2).
2. Analysis of contigs and identification of virus in figwort
In total, there were 133,962,402 raw reads with a GC content of 43.9% and Q30 of 94.61%. After trimming, 131,966,060 high-quality reads were obtained, which were assembled into 310,463 transcripts. The maximum and minimum contig lengths were 14,829 and 201 bp, respectively. The average contig length was 468.41 bp.
The NCBI BLAST search results of the assembled contigs revealed that one contig matched the previously reported the genome of the BWYV isolate LS (GenBank Accession No. KM076647), with the highest identity (97%). Detailed information on the contig is presented in Table 3.
3. Detection and identification of BWYV
RT-PCR amplification was performed to confirm the contig and infection rate. The expected 477 bp band was obtained and 30% (15/50) was positive for BWYV. Fifteen BWYV-positive samples showed 97.33% to 99.7% identity to different BWYV isolates in NCBI GenBank. This isolate identified from a figwort plant was named BWYV ADHS isolate (BWYV-ADHS).
4. Genome properties
The complete genome of BWYV-ADHS was 5,878 nucleotides (nt) in length and contained seven ORFs. The 5' and 3' untranslated regions (UTRs) consisted of 28 nt and 332 nt, respectively.
The 5' UTR contained the conserved sequence motif ACAAAA, which has been reported in other poleroviruses (Mo et al., 2010; Krueger et al., 2013).
The first ORF, ORF 0 (nt position: 29 - 745; amino acid [aa] length: 238), encodes a P0 protein domain proposed to be involved in the silencing of host defense (Bortolamiol et al., 2007).
ORF1 (nt position: 150 - 2,057; aa length: 635) encodes P1 (putative polyprotein) with the S39 peptidase motif that generates three proteins during virus replication (Miller et al., 2002; Nickel et al., 2008).
ORF2 (nt position: 150 - 3,331; aa length: 1,060) encodes the P1-P2 fusion protein of viral RNA-dependent RNA polymerase (RdRp) by a -1 slippery ribosomal frameshift at 1,454 nt (Knierim et al., 2013).
ORF3a (nt position: 3,414 - 3,551; aa length: 45) is initiated at an AUA codon and encodes ORF3a protein, which is involved in viral long-distance movement (Smirnova et al., 2015). ORF3 (nt position: 3,532 - 4,140; aa length: 202) and ORF4 (nt position: 3,563 - 4,090; aa length: 175) encode a coat protein (CP; P3 protein) and a movement protein (MP; P4 protein), respectively (Huang et al., 2005).
ORF5 (nt position: 3,532 - 5,547; aa length: 671) encodes the P5 protein (P3-P5 fusion protein) by read-through of the P3 stop codon (nt position: 4,138 - 4,140), which has been proposed to be responsible for aphid-mediated transmission (Brault et al., 2005).
The complete genome sequence was deposited in NCBI GenBank under the accession number LC592344, and the predicted genome organization of the BWYV ADHS isolate is shown in Fig. 3.
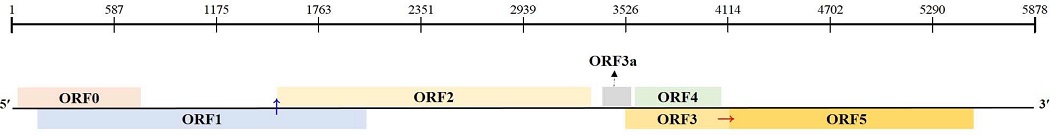
Schematic representation of genome characteristics of Beet western yellows virus (BWYV ADHS) isolate. Complete genome length; 5,878 nt, 5' and 3' UTR; 28 and 332 nt, ORF0 position; 29 - 745 nt, encodes P0 protein, ORF1; 150 - 2,057 nt, encodes P1 protein, ORF2; 150 - 333 nt, encodes P1-P2 fusion protein, ORF3a; 3,414 - 3,551 nt, encodes ORF3a protein, ORF3; 3,532 - 4,140 nt, encodes coat protein, ORF4; 3,563 - 4,090 nt, encodes movement protein, ORF5; 3,532 - 5,547 nt, encodes P3-P5 fusion protein. Blue arrow represents the position of the predicted -1 slippery ribosomal frameshift (1,454 nt). Red arrow indicates the predicted position of the leaky stop codon (4,140 nt).
5. Phylogenetic analysis and comparison identity
The phylogenetic analysis resolved the BWYV group into an Asian group (Korea, China, and Japan) and a non-Asian group (France and USA), except for the BWYV Shimizu isolate (Fig. 4).
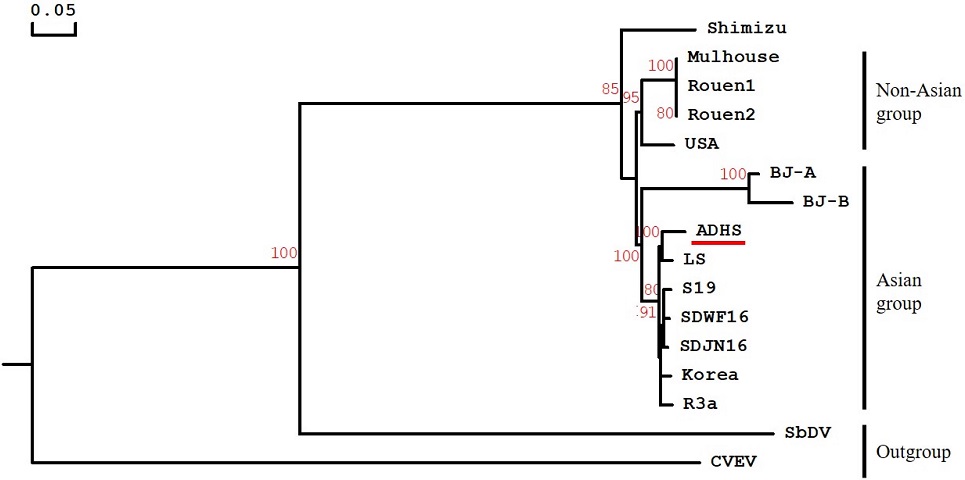
Phylogenetic tree of Beet western yellows virus (BWYV) ADHS isolate and previously reported isolates. The neighbor-joining tree was based on the complete nucleotide sequences. The red line indicates the strain obtained in this study. Bootstrap percentage values are based on 1,000 replications. Citrus vein enation virus (CVEV, Enamovirus genus) and Soybean dwarf virus (SbDV, Luteovirus genus) were used as outgroup. The corresponding sequence information and NCBI GenBank accession number for BWYV isolates used in the analysis are as follows; USA (NC004756), Korea (LC198684), Mulhouse (KU521324.1), Rouen1 (KU521325), Rouen2 (KU521326), LS (KM076647), ADHS (LC592344), SDWF16 (MK307780), SDJN16 (MK307779), R3a (LC428357), Shimizu (AB903032), S19 (LC428356), BJ-A (HM804471), BJ-B (HM804472), CVEV (NC021564), and SbDV (MT543032).
Phylogenetic trees based on the nucleotide complete genome confirmed that the BWYV ADHS isolate was most closely related to the LS isolate, to which it showed 96.1% nt sequence identity.
DISCUSSION
The recent advent of next generation sequencing (NGS) and bioinformatics has been successfully applied to detect the virus in various human tissues, plants, and soil microorganisms (Adams et al., 2009; Massart et al., 2014; Finley et al., 2015; Yuan et al., 2020). NGS is a powerful tool to identify the virus and its host (Villamor et al., 2019), and has been used in the discovery of more than 100 novel DNA and RNA plant viruses in recent years (Roossinck et al., 2015; Wu et al., 2015).
Novel or unreported viruses have been recorded from medicinal plants (Panax ginseng and Cnidium officinale) using NGS techniques in Korea (Yoo et al., 2015; Lee et al., 2020b). However, many studies using NGS focus only on sequence determination, and studies on biological properties, such as disease symptoms, are lacking. In particular, the analysis of damage to host plants caused by virus infection is very important, as it justifies the need to prevent and manage the threats from viruses.
For these reasons, we investigated the disease symptoms and compared growth characteristics of host plants. The main symptoms were dwarfism, mosaic pattern, and yellowing, and they were most frequently observed around the end of July in the open field.
As a result of RT-PCR diagnosis, a positive reaction for Beet western yellows virus was confirmed in 15 of 50 samples. Although all samples collected showed typical virus symptoms, no virus was detected in 35 samples. Therefore, additional studies of the causal agent that induces these symptoms in the 35 negative samples should be performed.
Previous research has shown that Polerovirus is transmitted by various aphid vectors (Knierim et al., 2014); for example, Myzus persicae and M. euphorbiae are capable of transmitting Potato leafroll virus (PLRV; type species in Polerovirus) (Srinivasan et al., 2008). M. persicae is a representative of heteroecious species that overwinter in the form of eggs in hosts, such as trees, and move to various field crops in the spring season.
Its fundatrix produces the alate, a winged young form, through parthenogenesis after overwintering, which then translocate to the summer host (Dixon et al., 1993). BWYV outbreaks in other plants and the presence of aphids have been reported in areas where figworts were collected (Kim and Kim, 2014; Kwon et al., 2018). Considering the life cycle of these aphids, it is thought that they were transmitted to figwort by aphids from the plant host grown in spring.
Growth characteristics of figwort plants were compared to measure the damage caused by viral infection. The plant length and root weight were decreased in the virus-infected figwort plants compared to the healthy plants. Dried roots of figwort plants are used as medicines (Kim et al., 2009b; 2012), and a decrease in root weight caused by the virus will result in serious economic losses.
To investigate the relationship of the isolated virus strain, a phylogenetic tree was constructed using different BWYV isolates. The results showed that BWYV-ADHS was closest to the LS isolate, which was identified in weeds from Korea (Kwon et al., 2016).
According to Kwon et al. (2016), Leonurus sibiricus may play an important role as a weed reservoir for pepper-infecting viruses such as BWYV. In Korea, peppers are cultivated nationwide not only for personal use, but also for sale. In a previous study on the occurrence pattern of viral disease in pepper, BWYV was the third most prevalent virus in open fields (Kwon et al., 2018).
Peppers are cultivated near the field where the figwort samples used in the present study were collected, and the BWYV ADHS isolate identified from the samples was expected to have a high correlation with peppers. However, the NCBI GenBank database has no complete genome seuqnece of a BWYV isolate from pepper in Korea. Therefore, an initial infection source that will include pepper and weeds should be investigated to verify the relationship of BWYV isolated from figwort plants.
To the best of our knowledge, this is the first report of BWYV in figwort plants worldwide. To minimize viral damage, it is important to recognize the damage caused by viral diseases and develop a control method that can be applied in figwort culture.
Acknowledgments
This work was supported by a grant (PJ012559082021) from the National Institute of Horticultural and Herbal Science, Rural Development Administration, Korea.
References
-
Adams IP, Glover RH, Monger WA, Mumford R, Jackeviciene E, Navalinskiene M, Samuitiene M and Boonham N. (2009). Next-generation sequencing and metagenomic analysis: A universal diagnostic tool in plant virology. Molecular Plant Pathology. 10:537-545.
[https://doi.org/10.1111/j.1364-3703.2009.00545.x]

-
Bortolamiol D, Pazhouhandeh M, Marrocco K, Genschik P and Ziegler-Graff V. (2007). The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Current Biology. 17:1615-1621.
[https://doi.org/10.1016/j.cub.2007.07.061]

-
Brault V, Périgon S, Reinbold C, Erdinger M, Scheidecker D, Herrbach E, Richards K and Ziegler-Graff V. (2005). The polerovirus minor capsid protein determines vector specificity and intestinal tropism in the aphid. Journal of Virology. 79:9685-9693.
[https://doi.org/10.1128/JVI.79.15.9685-9693.2005]

-
Candresse T, Marais A, Faure C, Lefebvre M, Lacombe T and Boursiquot JM. (2020). Complete genome sequence of a novel grapevine-infecting member of the genus Polerovirus, grapevine polerovirus 1. Archives of Virology. 165:1683-1685.
[https://doi.org/10.1007/s00705-020-04640-4]

-
Cheng PW, Ng LT, Chiang LC and Lin CC. (2006). Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clinical and Experimental Pharmacology and Physiology. 33:612-616.
[https://doi.org/10.1111/j.1440-1681.2006.04415.x]

-
Dixon AFG, Horth S and Kindlmann P. (1993). Migration in insects: Cost and strategies. Journal of Animal Ecology. 62:182-190.
[https://doi.org/10.2307/5492]

-
Domier LL. (2012). Family Luteoviridae. In King AMQ. et al. (eds.). Virus taxonomy: Ninth report of the International Committee on the Taxonomy of Viruses. Elsevier Academic Press. Amsterdam, Nederland. p.1045-1053.
[https://doi.org/10.1016/B978-0-12-384684-6.00090-2]

-
Finley SJ, Benbow ME and Javan GT. (2015). Potential applications of soil microbial ecology and next-generation sequencing in criminal investigations. Applied Soil Ecology. 88:69-78.
[https://doi.org/10.1016/j.apsoil.2015.01.001]

-
Huang LF, Naylor M, Pallett DW, Reeves J, Cooper JI and Wang H. (2005). The complete genome sequence, organization and affinities of carrot red leaf virus. Archives of Virology. 150:1845-1855.
[https://doi.org/10.1007/s00705-005-0537-6]

-
Kim ES and Kim YG. (2014). A report on mixed occurrence of tobacco whitefly(Bemisia tabaci) biotypes B and Q in Oriental melon farms in Kyungpook province, Korea. Korean Journal of Applied Entomology. 53:465-472.
[https://doi.org/10.5656/KSAE.2014.09.0.038]

-
Kim JK, Kim YH, Lee HH, Lim SS and Park KW. (2012). Effect of Scrophularia buergeriana extract on the degranulation of mast cells and ear swelling induced by dinitrofluorobenzene in mice. Inflammation. 35:183-191.
[https://doi.org/10.1007/s10753-011-9304-x]

- Kim JW, Lee SE, Lee JH, Min SJ, Kim TH, Lyu YS and Kang HW. (2009a). Study of Anti-Alzheimer activities from Scrophularia buergeriana water extract by alzheimer's protein APP-transgenic fly. Journal of Oriental Neuropsychiatry. 20:121-131.
-
Kim SJ, Park JS, Myung NY, Moon PD, Choi IY, An HJ, Kim NH, Na HJ, Kim DH, Kim MC, An NH, Kim IK, Lee JY, Jeong HJ, Um JY, Kim HM and Hong SH. (2009b). Scrophularia buergeriana regulates cytokine production in vitro. Immunopharmacology and Immunotoxicology. 31:246-252.
[https://doi.org/10.1080/08923970802432048]

-
Knierim D, Tsai WS, Deng TC, Green SK and Kenyon L. (2013). Full-length genome sequences of four polerovirus isolates infecting cucurbits in Taiwan determined from total RNA extracted from field samples. Plant Pathology. 62:633-641.
[https://doi.org/10.1111/j.1365-3059.2012.02653.x]

-
Knierim D, Tsai WS, Maiss E and Kenyon L. (2014). Molecular diversity of poleroviruses infecting cucurbit crops in four countries reveals the presence of members of six distinct species. Archives of Virology. 159:1459-1465.
[https://doi.org/10.1007/s00705-013-1939-5]

-
Krueger EN, Beckett RJ, Gray SM and Miller WA. (2013). The complete nucleotide sequence of the genome of Barley yellow dwarf virus-RMV reveals it to be a new Polerovirus distantly related to other yellow dwarf viruses. Frontiers in Microbiology. 4:205. https://www.frontiersin.org/articles/10.3389/fmicb.2013.00205/full, (cited by 2021 Feb 12).
[https://doi.org/10.3389/fmicb.2013.00205]

-
Kwon SJ, Cho IS, Yoon JY and Chung BN. (2018). Incidence and occurrence pattern of viruses on peppers growing in fields in Korea. Research in Plant Disease. 24:66-74.
[https://doi.org/10.5423/RPD.2018.24.1.66]

-
Kwon SJ, Choi GS, Yoon JY, Seo JK and Choi HS. (2016). Identification of Leonurus sibiricus as a weed reservoir for three pepper-infecting viruses. Plant Pathology Journal. 32:65-69.
[https://doi.org/10.5423/PPJ.NT.07.2015.0138]

-
Lee DH, Kim JK, Han JS, Lee JH, Lee B and Park CY. (2020a). Detection, isolation, and characterization of the cucumber mosaic virus in Pseudostellaria heterophylla from Korea. Journal of Plant Biotechnology. 47:150-156.
[https://doi.org/10.5010/JPB.2020.47.2.150]

-
Lee HK, Kim SY, Yang HJ, Lee D, Kwon B, Lee DY, Oh JH and Lee SH. (2020b). The detection of plant viruses in Korean ginseng(Panax ginseng) through RNA sequencing. Plant Pathology Journal. 36:643-650.
[https://doi.org/10.5423/PPJ.NT.07.2020.0137]

-
Li Y, Jiang S, Gao W and Zhu D. (2000). Phenylpropanoid glycosides from Scrophularia ningpoensis. Phytochemistry. 54:923-925.
[https://doi.org/10.1016/S0031-9422(00)00171-0]

-
Massart S, Olmos A, Jijakli H and Candresse T. (2014). Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Research. 188:90-96.
[https://doi.org/10.1016/j.virusres.2014.03.029]

-
Miller WA, Liu S and Beckett R. (2002). Barley yellow dwarf virus: Luteoviridae or Tombusviridae?. Molecular Plant Pathology. 3:177-183.
[https://doi.org/10.1046/j.1364-3703.2002.00112.x]

-
Mo X, Chen Z and Chen J. (2010). Complete nucleotide sequence and genome organization of a Chinese isolate of Tobacco vein distorting virus. Virus Genes. 41:425-431.
[https://doi.org/10.1007/s11262-010-0524-1]

-
Nickel H, Kawchuk L, Twyman RM, Zimmermann S, Junghans H, Winter S, Fischer R and Prüfer D. (2008). Plantibody-mediated inhibition of the Potato leafroll virus P1 protein reduces virus accumulation. Virus Research. 136:140-145.
[https://doi.org/10.1016/j.virusres.2008.05.001]

-
Pagán I and Holmes EC. (2010). Long-term evolution of the Luteoviridae: Time scale and mode of virus speciation. Journal of Virology. 84:6177-6187.
[https://doi.org/10.1128/JVI.02160-09]

-
Park CY, Baek DS, Oh JH, Choi JY, Bae DH, Kim JS, Jang GH and Lee SH. (2016). Survey of the incidence of viral infections in Calanthe spp. and characterization of a GW isolate of cymbidium mosaic virus in Korea. Research in Plant Disease. 22:65-71.
[https://doi.org/10.5423/RPD.2016.22.2.65]

-
Park CY, Kim JS, Lee HK, Oh JH, Lim SM, Moon JS and Lee SH. (2018). Beet western yellows virus (BWYV): Aspect of outbreak and survey, and first complete genome sequence of a Korea isolate of BWYV. Research in Plant Disease. 24:276-284.
[https://doi.org/10.5423/RPD.2018.24.4.276]

-
Park SU, Chae YA and Facchini PJ. (2003). Genetic transformation of the figwort, Scrophularia buergeriana Miq., an Oriental medicinal plant. Plant Cell Reports. 21:1194-1198.
[https://doi.org/10.1007/s00299-003-0639-0]

-
Peng B, Kang B, Wu H, Liu L, Liu L, Fei Z, Hong N and Gu Q. (2019). Detection and genome characterization of a novel member of the genus Polerovirus from zucchini(Cucurbita pepo) in China. Archives of Virology. 164:2187-2191.
[https://doi.org/10.1007/s00705-019-04217-w]

-
Reinbold C, Gildow FE, Herrbach E, Ziegler-Graff V, Goncalves MC, Van Den Heuvel JFJM and Brault V. (2001). Studies on the role of the minor capsid protein in transport of Beet western yellows virus through Myzus persicae. Journal of General Virology. 82:1995-2007.
[https://doi.org/10.1099/0022-1317-82-8-1995]

-
Roossinck MJ, Martin DP and Roumagnac P. (2015). Plant virus metagenomics: Advances in virus discovery. Phytopathology. 105:716-727.
[https://doi.org/10.1094/PHYTO-12-14-0356-RVW]

-
Scheunert A and Heubl G. (2011). Phylogenetic relationships among new world Scrophularia L.(Scrophulariaceae): New insights inferred from DNA sequence data. Plant Systematics and Evolution. 291:69-89.
[https://doi.org/10.1007/s00606-010-0369-z]

-
Smirnova E, Firth AE, Miller WA, Scheidecker D, Brault V, Reinbold C, Rakotondrafara AM, Chung BYW and Ziegler-Graff V. (2015). Discovery of a small non-AUG-initiated ORF in poleroviruses and luteoviruses that is required for long-distance movement. PLoS Pathogens. 11:e1004868. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1004868, (cited by 2021 Feb 16).
[https://doi.org/10.1371/journal.ppat.1004868]

-
Srinivasan R, Alvarez JM, Bosque-Pérez NA, Eigenbrode SD and Novy RG. (2008). Effect of an alternate weed host, hairy nightshade, Solanum sarrachoides, on the biology of the two most important Potato leafroll virus(Luteoviridae: Polerovirus) vectors, Myzus persicae and Macrosiphum euphorbiae (Aphididae: Homoptera). Environmental Entomology. 37:592-600.
[https://doi.org/10.1093/ee/37.2.592]

-
Villamor DEV, Ho T, Al Rwahnih M, Martin RR and Tzanetakis IE. (2019). High throughput sequencing for plant virus detection and discovery. Phytopathology. 109:716-725.
[https://doi.org/10.1094/PHYTO-07-18-0257-RVW]

-
Wu Q, Ding S, Zhang Y and Zhu S. (2015). Identification of viruses and viroids by next-generation sequencing and homology-dependent and homology-independent algorithms. Annual Review of Phytopathology. 53:425-444.
[https://doi.org/10.1146/annurev-phyto-080614-120030]

-
Yoo RH, Zhao F, Lim SM, Igori D, Kim SM, An TJ, Lee SH and Moon JS. (2015). The complete genome sequences of two isolates of cnidium vein yellowing virus, a tentative new member of the family Secoviridae. Archives of Virology. 160:2911-2914.
[https://doi.org/10.1007/s00705-015-2557-1]

-
Yuan Z, Ye X, Zhu L, Zhang N, An Z and Zheng WJ. (2020). Virome assembly and annotation in brain tissue based on next-generation sequencing. Cancer Medicine. 9:6776-6790.
[https://doi.org/10.1002/cam4.3325]
