Anti-inflammatory Activity of Philadelphus schrenkii Methanol Extract in Lipopolysaccharide-stimulated RAW 264.7 Cells
This is an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Philadelphus schrenkii Rupr. is a widespread in East Asia and has historically been used as a medicinal herb in Korea to reduce heat, swelling, and pain. This study investigated the anti-inflammatory effects of P. schrenkii methanol extract (PSE) on lipopolysaccharide-stimulated RAW 264.7 macrophage cells.
PSE inhibited the expression of nitric oxide, prostaglandin E2, inducible nitric oxide synthase, and cyclooxygenase-2 at concentrations that did not affect cell viability as determined by the tetrazolium salt (WST-1) assay. The quantitative reverse transcription polymerase chain reaction and enzyme-linked immunosorbent assay measured that pro-inflammatory cytokines, including tumor necrosis factor-α, interleukin (IL)-1β, IL-6, and monocyte chemoattracted protein-1, were decreased in PSE-treated groups. Western blotting confirmed that PSE inhibited the phosphorylation of nuclear factor kappa B and mitogen-activated protein kinases. Gas chromatography-mass spectrometry analysis revealed the presence of active anti-inflammatory compounds in PSE.
This study demonstrated that PSE has potent anti-inflammatory activity in vitro and provides a reasonable basis for the traditional use of P. schrenkii in treating inflammation-related diseases.
Keywords:
Philadelphus schrenkii Rupr., Anti-inflammatory, Mitogen Activated Protein Kinase, Nuclear Factor Kappa B, RAW 264.7 CellsINTRODUCTION
Inflammation is a necessary defense mechanism for living organisms to protect themselves against tissue injuries, infections, and harmful external stimuli. Inflammation can be triggered by damage-associated molecular patterns, foreign nucleic acids, or pathogen-associated molecular patterns such as lipopolysaccharide (LPS) (Newton and Dixit, 2012). During the inflammatory response, macrophages receive stimuli such as LPS into the cells and produce nitric oxide (NO), prostaglandin E2 (PGE2), and pro-inflammatory cytokines (Yang et al., 2012; Kim et al., 2019). And the production of these inflammatory factors requires the activation of the nuclear factor-κB (NF-κB) and mitogen-activated protein kinase (MAPK) signaling pathways (Liu et al., 2018; Cao et al., 2021).
Although inflammation is generally a beneficial mechanism, chronic inflammation can lead to the development of various diseases, including rheumatoid arthritis, metabolic syndromes, and cancer (Chippada et al., 2011; Kim et al., 2014; Dong et al., 2017). Aspirin, metformin, and diclofenac, which are inhibitors of inflammation-related enzymes such as inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2), are anti-inflammatory agents that can help prevent the development of various degenerative diseases (Munn, 2017; Chen et al., 2018; Nore et al., 2020; Chakrabarti and Mukherjee, 2021). Therefore, reducing inflammation is essential to preventing a number of degenerative diseases.
Approximately 49% of newly approved medicines worldwide in the last 39 years (1981 - 2019) were either natural products or derived from natural sources. Notably, 25% of pharmaceutical products are derived from plants. Natural products are an important source of new drug development because of their chemical and structural variety, which often results in better bioactivity than synthesized molecules (Newman and Cragg, 2020). For these reasons, numerous studies have recently been reported on research into natural anti-inflammatory agents (Nunes et al., 2020).
The genus Philadelphus L., which has 75 species, is found in East Asia, North and South America, and the Himalayas (Park et al., 2005). The flowers and fruits of P. schrenkii are traditionally used as herbal remedies in South Korea to reduce fever, swelling, and pain (Xiao, 1989). The Hydrangeaceae family, to which Philadelphus schrenkii Rupr. belongs, has been reported to have antibacterial, anti-inflammatory, anti-obesity, antidiabetic, and hepatoprotective activities (Nakamura et al., 2011; Shi et al., 2015; Myung et al., 2020; Sung et al., 2023).
In the case of P. schrenkii, it has antibacterial and antioxidant activity, as reported in a study by Shin et al. (1997), Lee et al. (1998), and Kim et al. (2022). In particular, the NO inhibitory ability was confirmed to analyze the correlation between the phenolic acid content and the physiological activity of P. schrenkii extract in the study of Kim et al. (2021). However, the molecular mechanism of the anti-inflammatory activity of P. schrenkii has not been studied to date.
Therefore, this study aims to evaluate the anti-inflammatory activity of P. schrenkii methanol extract (PSE) by measuring the inhibitory effect of NO production in LPS-activated RAW 264.7 cells. Additionally, the study seeks to understand the anti-inflammatory mechanism of PSE by examining the expression of various proteins and inflammatory factors, including NF-κB.
MATERIALS AND METHODS
1. Preparation of Plant Extract
The P. schrenkii methanol extract (KPM033-007) used in this research was obtained from the Natural Product Central Bank at the Korea Research Institute of Bioscience and Biotechnology (Daejeon, Korea). The plant was collected from Inje-gun, Gangwon-do, Korea in 2008. A voucher specimen (KRIB 0017867) is kept in the herbarium of the Korea Research Institute of Bioscience and Biotechnology.
The plant (69 g) dried in the shade and powdered was added to 1 ℓ of methyl alcohol 99.9% (HPLC grade) and extracted through 30 cycles (40 ㎑, 1,500 W, 15 min. Ultrasonication - 120 min. standing per cycle) at room temperature using an ultrasonic extractor (SDN-900H, SD-ULTRASONIC Co., Ltd, Seoul, Korea). After filtration (Qualitative Filter No.100, HYUNDAI MICRO Co., Ltd, Seoul, Korea) and drying under reduced pressure, P. schrenkii extract (6.2 g) was obtained.
The extract was dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich Co., Saint Louis, MO, USA) at a concentration of 100 ㎎/㎖ and kept at 4℃.
2. Cell Culture
Murine macrophage RAW 264.7 cells (American Type Culture Collection, Manassas, VA, USA) were grown in Dulbecco’s modified Eagle’s medium (DMEM, Corning™, Corning, NY, USA) with 10% heat-inactivated fetal bovine serum (FBS, Corning™, Corning, NY, USA) and a 1% mixture of penicillin and streptomycin (Corning™, Corning, NY, USA). Cells were incubated at 37℃ under humidified conditions with 5% CO2.
3. Cell Viability assay
To determine the cytotoxicity of PSE, RAW 264.7 cells were seeded in a 96-well cell culture plate with a density of 1 × 104 cells per well and incubated for 24 h. The cells were treated with various concentrations (50, 100, 150, and 200 ㎍/㎖) of PSE for 24 h.
After treatment, the medium was replaced with a fresh one, and 10 ㎕ of EZ-cytox Cell Viability Assay Solution WST-1 (Daeil Lab Service, Seoul, Korea) was added to each well. Then, the cells were further incubated for 90 minutes at 37℃. The absorbance was measured at 460 ㎚ using a VersaMax™ microplate reader (Molecular Devices, San Jose, CA, USA).
4. Nitric oxide (NO) assay
Following preincubation of RAW 264.7 cells (5 × 104 cells/well) in 24-well cell culture plates for 22 h, the medium was replaced with a fresh medium.
Then, various concentrations (50, 100, 150, and 200 ㎍/㎖) of PSE were treated for 2 h. Afterward, 1 ㎍/㎖ of LPS derived from Escherichia coli O111:B4 (Sigma-Aldrich Co., Saint Louis, MO, USA) was added to each well and further incubated for 24 h.
To analyze NO production, 100 ㎕ of the culture supernatant was transferred to 96-well plates, and the same amount of Griess reagent (Sigma-Aldrich Co., Saint Louis, MO, USA) was added. The plates were incubated for 15 min at room temperature in dark conditions.
The absorbance was determined by the VersaMax™ microplate reader (Molecular Devices, San Jose, CA, USA) at a wavelength of 540 ㎚. A standard curve of NaNO2 was used for the calculation of the nitrite level.
5. Prostaglandin E2 (PGE2) Quantification Assay
RAW 264.7 cells were cultured in 24-well plates for 24 h, and then treated with 100 and 200 ㎍/㎖ of PSE for 2 h. Afterward, the culture supernatant was collected to quantify the amount of PGE2 secreted by RAW 264.7 cells that were activated by exposure to 1 ㎍/㎖ of LPS for 22 h.
The investigation of PGE2 production levels was performed following the protocol provided by the manufacturer of the PGE2 Parameter Assay Kit (R&D Systems Inc., Minneapolis, MN, USA). The absorbance was measured at 450 ㎚ using the VersaMax™ microplate reader (Molecular Devices, San Jose, CA, USA).
6. Enzyme-Linked Immunosorbent Assay (ELISA)
The culture supernatant was collected after treating the RAW 264.7 cells with PSE for 2 h and stimulating them with LPS for 22 h.
The levels of tumor necrosis factor-alpha (TNF-α) and monocyte chemoattractant protein-1 (MCP-1) secreted by LPS-stimulated RAW 264.7 cells were quantified using the TNF-α Mouse ELISA Kit and the MCP-1 Mouse ELISA Kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol.
Absorbance measurements were taken using the VersaMax™ microplate reader (Molecular Devices, San Jose, CA, USA) at a wavelength of 450 ㎚.
7. Reverse Transcription Quantitative Real-time Polymerase Chain Reaction (RT-qPCR)
The mRNA expression levels of inflammatory proteins, including iNOS and COX-2, along with inflammatory cytokines such as TNF-α, interleukin (IL)-1β, IL-6, and MCP-1, were investigated by RT-qPCR. The PSE was incubated with RAW 264.7 cells for 2 h, followed by the addition of LPS for 22 h.
The cells were then harvested, and total mRNA was isolated using the standard protocol of the RNeasy® Plus Mini Kit (Qiagen, Hilden, Germany). The isolated mRNA was assessed for purity and quantified at 260 ㎚ and 280 ㎚ wavelengths using a NanoDrop™ 2000 Spectrophotometer (Thermo Fisher Scientific™, Waltham, MA, USA). RT-PCR was performed using AccuPower® CycleScript™ RT PreMix (dT20) (Bioneer, Daejeon, Korea) to synthesize 1 ㎍ of mRNA into cDNA. The primer sequences utilized in qPCR are listed in Table 1.

Primer sequences of mRNA expression levels of inflammation-related genes by treatment of P. schrenkii methanol extract (PSE) in quantitative real-time polymerase chain reaction.
The qPCR reaction mixture was prepared by adding 10 ㎕ of AccuPower® 2x GreenStar™ qPCR Master Mix (Bioneer) containing SYBR green dye, 1 ㎕ of forward primer (10 pmol), 1 ㎕ of reverse primer (10 pmol), 1 ㎕ of synthesized cDNA, and distilled water to a final volume of 20 ㎕.
qPCR was performed using the QuantStudio™ 6 Flex System (Thermo Fisher Scientific™). The qPCR conditions are as follows: pre-denaturation for 10 min at 95℃, denaturation for 15 sec at 95℃, annealing for 30 sec at 60℃, extension for 30 sec at 72℃, repeated for 40 cycles, and final extension for 5 min at 72℃. After completion of the reaction, the qPCR reaction mixture was subjected to denaturation at 95℃ for 15 sec, followed by annealing at 60℃ for 1 minute. The fluorescence signal for melt curve analysis was measured as the temperature was increased from 60℃ to 95℃ at a rate of 0.05℃/s.
The amount of total mRNA was normalized using GAPDH, a housekeeping gene. The comparative Ct (ΔΔCt) approach was then used to calculate the relative mRNA expression levels of each target gene.
8. Western Blot Analysis
RAW 264.7 cells (1.5 × 106 cells/㎖) were cultured for 24 h and pre-treated with PSE (100 and 200 ㎍/㎖) for 2 h. After adding LPS (1 ㎍/㎖), the cells were incubated for 30 min or 22 h.
Whole-cell lysates were prepared by washing and harvesting the cells with ice-cold phosphate-buffered saline (PBS, Biosesang, Seongnam, Korea), and lysing them with ice-cold PRO-PREP™ Protein Extraction Solution (iNtRON Biotechnology, Seongnam, Korea) in ice for 30 min.
The supernatant was obtained by performing centrifugation at 13,200 rpm for 20 min at 4℃. To prepare nuclear and cytoplasmic protein extracts, RAW 264.7 cells were treated with PSE for 2 h. The cells were then collected 30 min after LPS stimulation.
The nuclear and cytoplasmic fractions were separated and extracted following the manufacturer's instructions for NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific™, Waltham, MA, USA).
The protein concentration was quantified using the Bradford reagent (Biosesang) and calculated with the BSA standard. An equal concentration of protein was prepared with Laemmli Sample Buffer (Bio-Rad, Hercules, CA, USA) and then boiled for 5 min.
Protein samples were separated on 12% sodium dodecyl sulfate-polyacrylamide gels and subsequently transferred onto nitrocellulose membranes (PALL Life Sciences, Port Washington, NY, USA). The membrane was blocked for 1 h in PBST (PBS with 0.1% Tween 20) containing 5% skim milk (BioShop, Ontario, Canada). After blocking, the membrane was incubated with the primary antibodies overnight at 4℃.
The primary antibodies used in this experiment included iNOS, COX-2, IKKα, IKKβ, p-IKKα/β (Ser176/180), IκBα, p-IκBα (Ser32), NF-κB, p-NF-κB p65 (Ser536), ERK 1/2, p-ERK 1/2 (Thr202/Tyr204), SAPK/JNK, p-SAPK/JNK (Thr183/Tyr185), p38, p-p38 (Thr180/Tyr182), β-actin, and Histone H3 (Cell Signaling Technology Inc., Danvers, MA, USA).
The membrane was washed three times with PBST buffer and then incubated with a horseradish peroxidase-conjugated secondary antibody (either anti-rabbit IgG or anti-mouse IgG, Cell Signaling Technology Inc., Denver, MA, USA) for 1 h at room temperature.
Afterward, the membrane was rewashed three times. Enhanced chemiluminescence (ECL) solution (AbFrontier, Seoul, Korea) was used for protein development. The signals were detected with iBright™ CL1500 Imaging System (Thermo Fisher Scientific™, Waltham, MA, USA).
9. Gas Chromatography-Mass Spectrometry (GC-MS) Analysis
The compounds of PSE were analyzed using a GCMS-QP2010 Ultra (Shimadzu, Kyoto, Japan).
The PSE sample solution dissolved in methanol was injected at a flow rate of 1 ㎖/min and a split ratio of 10 : 1 with splitless injection mode into the Agilent DB-5ms Ultra Inert Columns (30 m × 0.25 ㎜ × 0.25 ㎛) (Agilent Technologies, Santa Clara, CA, USA) using helium as a carrier gas. The injection temperature was 250℃, and the temperature program was set to 2 min at 50℃, 5 ℃/min from 50℃ to 250℃, 10 ℃/min from 250℃ to 320℃, and hold for 11 min at 320℃.
MS analysis conditions were electron ionization at 70 eV, an ion source temperature of 200℃, a solvent cut time of 5 min, and a scanning interval of 0.3 sec at a 40 to 600 m/z scan range.
10. Statistical Analysis
Data from all experiments are reported as mean values ± standard deviation (SD) based on at least three replicates. Statistical analysis was conducted using GraphPad Prism 9 software (GraphPad Software, San Diego, CA, USA).
Statistical significance was evaluated with One-way ANOVA, followed by Dunnett's Multiple Comparison Test to analyze the differences between each group. When p < 0.05 was achieved, differences were considered statistically significant.
RESULTS AND DISCUSSION
1. Effects of P. schrenkii methanol extract (PSE) on cell viability
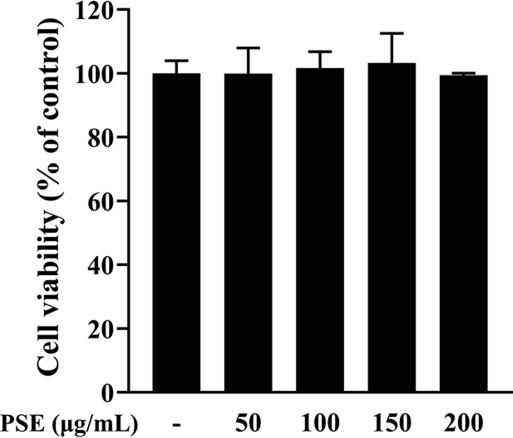
To investigate the cytotoxic effect, the PSE was treated at various concentrations (50, 100, 150, and 200 ㎍/㎖) on RAW 264.7 cells and measured using the WST-1 assay.
RAW 264.7 cells are mice-derived macrophages that produce a higher amount of inflammatory mediators, such as NO and PGE2, and promote phagocytosis when stimulated by LPS (Taciak et al., 2018). Therefore, the discovery of an agent that can regulate macrophage activation and inhibit the secretion of inflammatory mediators is considered an effective way to develop a new anti-inflammatory drug (Konishi et al., 2008; Marasinghe et al., 2022; Seo et al., 2022).
Cell viability assay results of PSE did not show any cytotoxic effects at concentrations up to 200 ㎍/㎖ compared to the control (Fig. 1). Therefore, the PSE was used up to a concentration of 200 ㎍/㎖ in the study.
2. Effects of P. schrenkii methanol extract (PSE) on the inflammatory response
In order to investigate the anti-inflammatory capacity of PSE, the NO and PGE2 production in LPS-induced RAW 264.7 cells were examined using a non-toxic dose range of the PSE. NO is a signaling molecule that plays vital roles in several human physiological systems (Vuolteenaho et al., 2007).
NO is produced when inflammation is induced by various factors, such as stimulation of exposure, and is an essential molecule for the immune response as it has the ability to eliminate bacteria and viruses (Wink et al., 2011). However, excessive production of NO can act as a pro-inflammatory mediator and cause tissue damage and inflammation. Therefore, inhibition of NO production is crucial in modulating the inflammatory response (Sharma et al., 2007).
PGE2 causes edema and redness by dilating arteries and increasing vascular permeability in inflammation. It also affects the nervous system to cause pain (Funk, 2001). Therefore, the anti-inflammatory effect of PSE was confirmed by measuring the production of NO and PGE2, which are inflammatory mediators.
The results showed that, compared to the control group treated only with LPS, the levels of NO and PGE2 production were remarkably reduced to 19.1% and 15.04%, respectively, in the group treated with PSE at a concentration of 200 ㎍/㎖ (Fig. 2A and B). These results indicated that the PSE could suppress inflammatory responses through the inhibition of NO and PGE2 production.
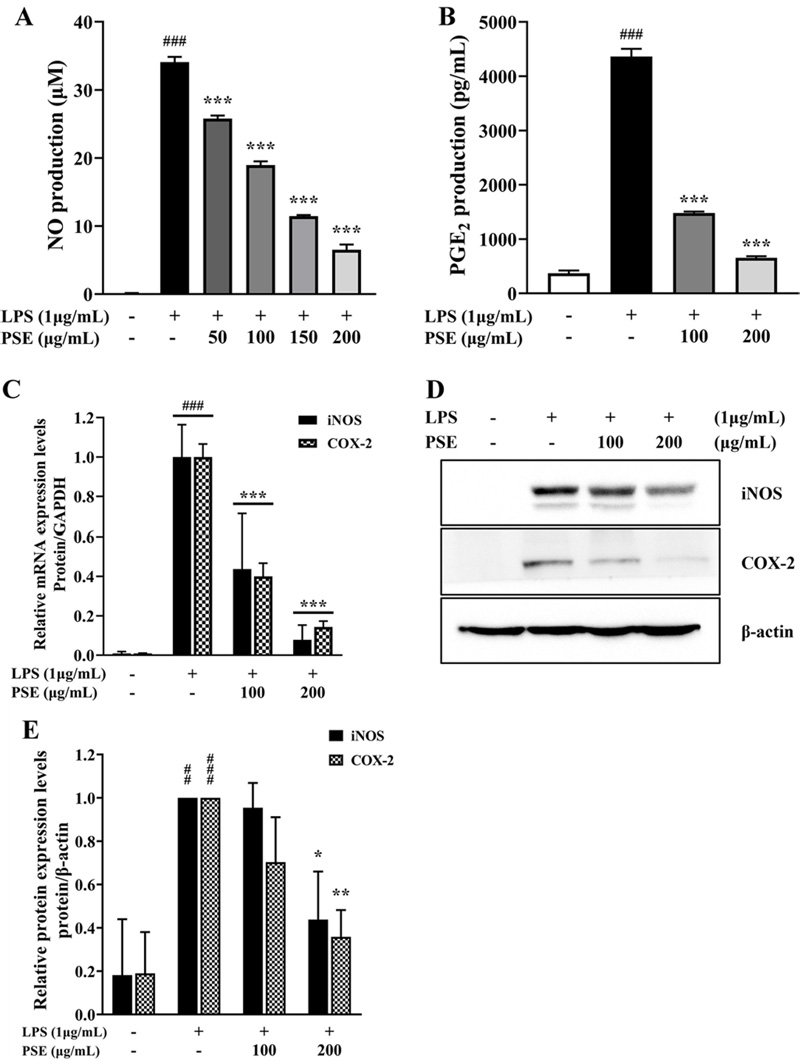
Effects of P. schrenkii methanol extract (PSE) on the inflammatory response in LPS-treated RAW 264.7 cells.(A) The NO production was measured in the cell supernatant using the Griess reagent. (B) The PGE2 synthesis level was analyzed using the PGE2 assay kit. (C) The mRNA expression levels of iNOS and COX-2 in LPS-activated RAW 264.7 cells were determined using RT-qPCR. The results were normalized to the Ct value of GAPDH and analyzed by the comparative Ct method. (D) The western blot assay result, and protein level of iNOS and COX-2 protein expression in whole-cell lysates. β-actin was used as a loading control. (E) The intensity of iNOS and COX-2 proteins was quantified relative to the β-actin using ImageJ. Data represent the means ± SD (n = 3). ##p < 0.01 and ###p < 0.001 compared to the non-treated group, *p < 0.05 and **p < 0.01 compared to the LPS-only treated cells.
RT-qPCR and western blot analysis were performed to verify the protein and mRNA expression levels of two major inflammatory enzymes, iNOS and COX-2, in order to examine the effects of PSE on NO production further. The iNOS is one of the isoforms of nitric oxide synthase that synthesizes L-arginine into NO. The expression of NOS is induced by LPS or cytokines such as IL-1 and TNF-α (Miyasaka and Hirata, 1997; Habib and Ali, 2011).
Similarly, COX is also considered a key enzyme in inflammatory reactions and has two isoforms, COX-1 and COX-2. After being stimulated by pro-inflammatory factors, COX-2 produces one of the major inflammatory mediators, particularly PGE2, by converting arachidonic acid (Simon, 1999).
Results indicated that, compared to the LPS-only treatment group, the mRNA expression of iNOS and COX-2 in the PSE treatment group was suppressed by up to 7.83% and 14.37%, respectively (Fig. 2C).
Furthermore, protein expression was suppressed by up to 43.84% for iNOS and up to 35.84% for COX-2 when PSE was added (Fig. 2D and 2E). This indicated that the PSE treatment effectively suppressed both the mRNA and protein expressions of iNOS and COX-2. These results demonstrated that the PSE reduced NO and PGE2 production by preventing the expression of iNOS and COX-2 in LPS-induced RAW 264.7 cells.
3. Effects of P. schrenkii methanol extract (PSE) on the expression of pro-inflammatory cytokines
To evaluate the anti-inflammatory effects of PSE, the production and mRNA expression levels of inflammation-related cytokines, including TNF-α, IL-1β, IL-6, and MCP-1, were measured using ELISA and RT-qPCR.
TNF-α maintains the nuclear translocation of NF-κB, which activates the expression of inflammation-associated genes such as iNOS and COX-2 (Dinarello, 2000). MCP-1 is an inflammatory cytokine that is transcriptionally upregulated by NF-κB during the inflammation response and strongly attracts monocytes (Yadav et al., 2010).
In addition, the continuous release of pro-inflammatory cytokines can lead to the development of chronic inflammatory diseases. IL-1β acts as an important inflammatory mediator and causes inflammatory pain hypersensitivity. IL-6 plays an essential role in the transition from acute to chronic inflammation (Gabay, 2006).
As shown in Fig. 3A, in comparison to the LPS control, the PSE demonstrated inhibition of TNF-α and MCP-1 levels to 49.9% and 45.06%, respectively. The PSE also showed inhibitory activities against the mRNA expressions of inflammatory cytokines in LPS-induced RAW 264.7 cells. The mRNA expression levels of TNF-α, IL-1β, IL-6, and MCP-1 treated with the PSE of 200 ㎍/㎖ were considerably reduced to 44.94%, 8.94%, 2.54%, and 49.75%, respectively, compared to the group treated with LPS alone (Fig. 3B).
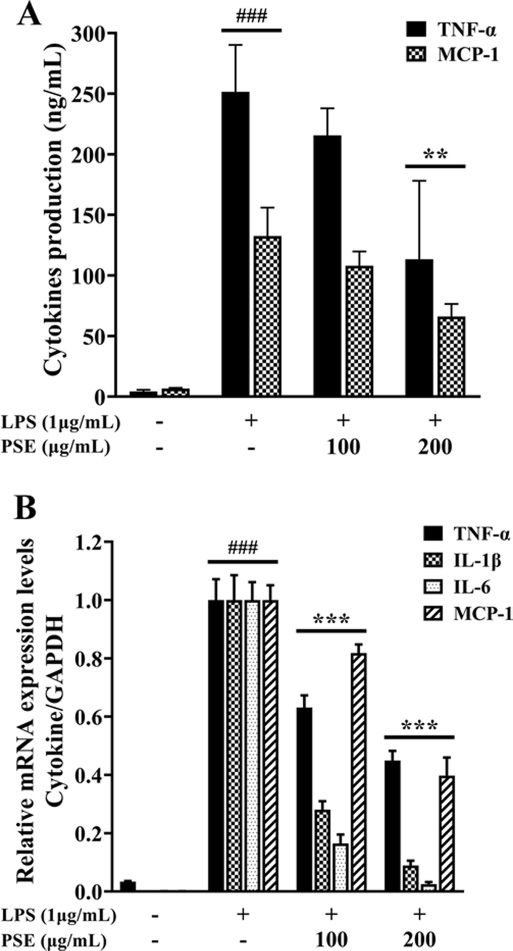
Effects of P. schrenkii methanol extract (PSE) on the production of pro-inflammatory cytokines.(A) The ELISA kits were used to measure the release levels of TNF-α and MCP-1 secreted into the supernatant. (B) The mRNA expression levels of TNF-α, IL-1β, IL-6, and MCP-1 were estimated using RT-qPCR. The results were normalized to the Ct value of GAPDH and analyzed by the comparative Ct method. Data represent the means ± SD (n = 3). ###p < 0.001 compared to the nontreated group, **p < 0.01 and ***p < 0.001 compared to the LPS-only treated cells.
These results suggested that PSE effectively prevented the development of chronic inflammation by reducing the synthesis and mRNA expression of pro-inflammatory cytokines.
4. Inhibitory effects of P. schrenkii methanol extract (PSE) on the NF-κB in LPS-stimulated RAW 264.7 cells
Western blot analysis was performed to determine the phosphorylation levels of proteins in the NF-κB and MAPK signaling pathways.
Inflammatory responses are known to be significantly regulated by the activation of the NF-κB and MAPK signaling pathways (Jayawardena et al., 2021). NF-κB is a crucial transcriptional regulatory factor that affects inflammation, cell proliferation, and apoptosis (Viatour et al., 2005). In the quiescent state, NF-κB exists in the cytoplasm as an inactive form combined with its suppressor, NF-κB inhibitor alpha (IκBα). When the cells are exposed to LPS, the IκB kinase (IKK) complex phosphorylates IκBα and causes the degradation of IκBα. As a result, NF-κB, liberated from IκBα, is activated and translocated to the nucleus (Jayawardena et al., 2021). Nuclear translocation of NF-κB initiates the transcription of genes that encode pro-inflammatory cytokines, including TNF-α, IL-1β, and IL-6, and inflammatory mediators, such as iNOS and COX-2 (Kim et al., 2018; Asanka Sanjeewa et al., 2019).
According to the results shown in Fig. 4A and 4C, PSE did not affect the total expression of IKKα/β and NF-κB. In addition, the phosphorylation levels of IKKα/β and NF-κB compared to their total expression levels were suppressed by 19.57% and 3.24%, respectively, in the 200 ㎍/㎖ PSE-treated group. In the LPS-only treated group, total IκBα protein expression was decreased; in the PSE-treated group, it was not downregulated. But PSE at 200 ㎍/㎖ reduced LPS-induced IκBα phosphorylation to 38.97%. Our results indicated that the expression of total IκBα was suppressed by LPS, and PSE reduced the LPS-induced phosphorylation of IκBα. This also means that PSE inhibits the phosphorylation of IKKα/β and IκBα, thereby interfering with the activation of NF-κB.
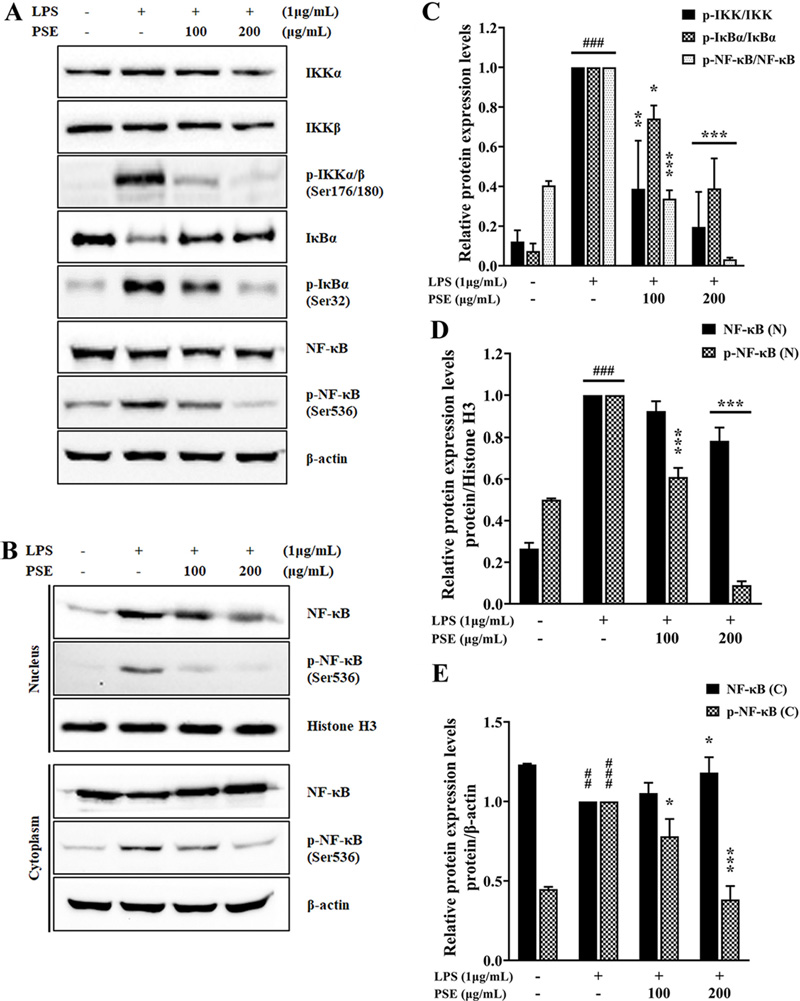
Effects of P. schrenkii methanol extract (PSE) on the NF-κB signaling pathway in LPS-induced RAW 264.7 cells.The expression levels of total and phosphorylated forms of (A) IKKα/β, IκBα, and NF-κB in whole-cell lysates, and (B) NF-κB in the nuclear and cytoplasmic fractions were determined by western blot analysis. β-actin and Histone H3 were used as loading controls. The relative protein expression levels of (C) p-IKK/IKK, p-IκBα/IκBα, and p-NF-κB/NF-κB, (D) NF-κB/Histone H3 and p-NF-κB/Histone H3 in nuclear fraction, (E) NF-κB/Histone H3 and p-NF-κB/Histone H3 in cytoplasmic fraction. The intensity of proteins was measured using ImageJ. Data represent the means ± SD (n = 3). ##p < 0.01 and ###p < 0.001 compared to the non-treated group, *p < 0.05, **p < 0.01, and ***p < 0.001 compared to the LPS-only treated cells.
Afterwards, we verified the translocation of NF-κB from the cytoplasm to the nucleus. Activated NF-κB is necessary for inducing responses to several immunological stimuli by binding to the promoter/enhancer regions of pro-inflammatory cytokine/ chemokine genes (Smale, 2011). In other words, targeting NF-κB signaling could be a useful therapeutic strategy for inflammatory diseases (Ben Neriah and Karin, 2011). Regulation of NF-κB signaling can be achieved through modulation of several steps between receptor activation and initiation of gene transcription, nuclear translocation, DNA binding, or interference with transcription initiation of NF-κB target genes (Ramadass et al., 2020).
As shown in Figures 4B and 4D, the expression of total and phosphorylated NF-κB in the nuclear fraction gradually reduced in the PSE-treated group. In the cytoplasmic fraction, NF-κB phosphorylation increased upon LPS stimulation and decreased with increasing PSE concentration (Fig. 4B and 4E). Conversely, although total cytoplasmic NF-κB was decreased by LPS, its expression was not decreased in the PSE-treated group. Thus, the result of inhibition of nuclear translocation of NF-κB in Fig. 4B indicates that PSE can be helpful in suppressing inflammation.
These results showed that PSE effectively interrupted the activation and nuclear translocation of NF-κB in LPS-activated RAW 264.7 cells by modulating IκBα and IKKα/β phosphorylation levels.
5. Effects of P. schrenkii methanol extract (PSE) on the MAPK pathway
The phosphorylation of MAPK was examined for further investigation into the anti-inflammatory activities of PSE. MAPK, which consists of extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK), and p38 MAPK, regulates biological activities such as cell growth, differentiation, and apoptosis. They are also involved in inflammation, innate immune systems, and cancer (Wang et al., 2023). MAPK proteins are activated by mitogenic stimuli, cytokines, various growth factors, and pro-inflammatory or stressful responses (Kim et al., 2005; Zhang et al., 2019). When MAPK proteins are phosphorylated by LPS stimulation, they can enhance the transcription of inflammatory genes by inducing the activation of downstream transcription factors, especially NF-κB (Kim et al., 2018).
In the case of the MAPK pathway, the phosphorylation levels of ERK, JNK, and p38 were reduced to 36.06%, 44.64%, and 57.75%, respectively, without significant alteration of total protein expressions in 200 ㎍/㎖ PSE-treated groups compared to those in the LPS-stimulated group (Fig. 5A and 5B). These results demonstrate that the anti-inflammatory activity of PSE results from inhibiting NF-κB activity by interfering with the phosphorylation of NF-κB upstream proteins, including the IKK complex, IκBα, and MAPK family.
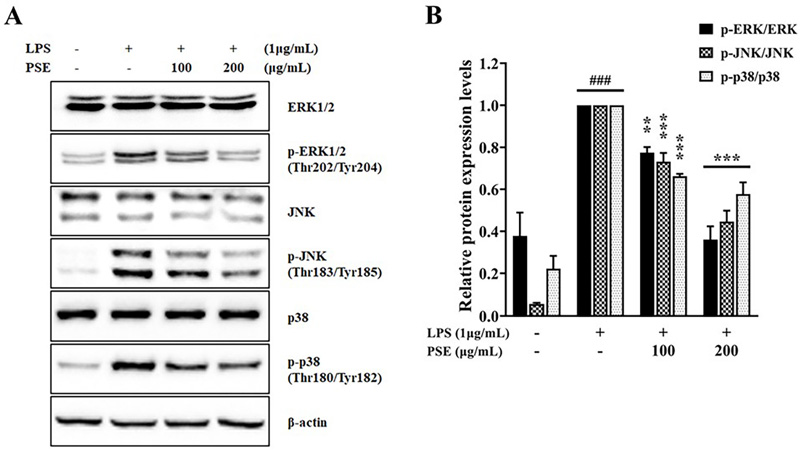
Effects of P. schrenkii methanol extract (PSE) on the MAPK signaling pathway in LPS-induced RAW 264.7 cells.The expression levels of total and phosphorylated forms of (A) ERK, JNK, and p38 were determined by western blot analysis in whole-cell lysates. β-actin was used as a loading control. The relative protein expression levels of (B) p-ERK/ERK, p-JNK/JNK, and p-p38/p38. The intensity of proteins was measured using ImageJ. Data represent the means ± SD (n = 3). ###p < 0.001 compared to the non-treated group, **p < 0.01 and ***p < 0.001 compared to the LPS-only treated cells.
6. GC-MS analysis of the P. schrenkii methanol extract (PSE)
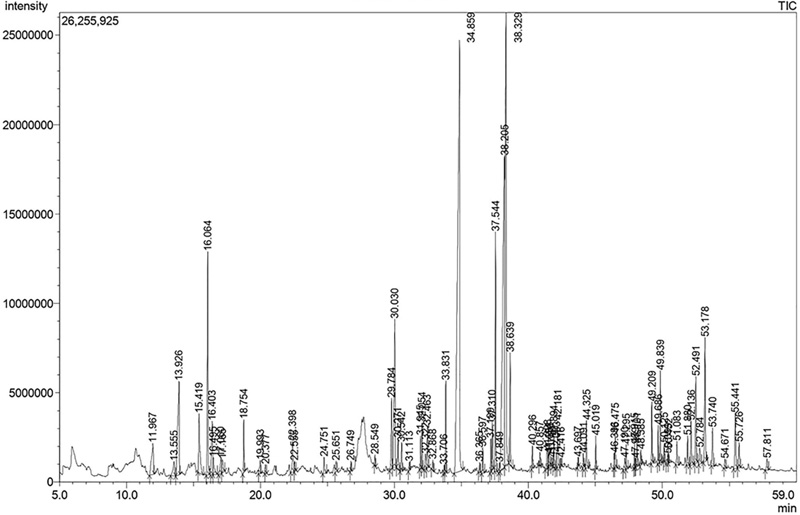
The ingredients of the PSE were investigated by GC-MS to identify the compounds that enable it to have an anti-inflammatory effect. A total of 79 compounds were detected in the GC-MS analysis of PSE (Fig. 6), and 38 compounds (their peak area accounted for 80.54% of the entire peak area of the chromatogram) were clarified as a result of comparing the mass spectrum of the compound with the National Institute of Standards and Technology (NIST)11 and Wiley9 mass spectral databases (Table 2).
An overview of the identified compounds is shown in Table 2, and 25 have been reported to have anti-inflammatory activity. These compounds accounted for 73.69% of the total peak area in the chromatogram.
Among the identified compositions, the most abundant compound in the PSE was n-Hexadecanoic acid (Retention time 34.859 min), well known as palmitic acid, accounting for 20.31%. n-hexadecanoic acid has anti-inflammatory activity by reducing the synthesis of inflammatory mediators such as NO, PGE2, TNF-α, IL-1β, and IL-6 (Aparna et al., 2012).
The second most abundant compound in PSE is 9,12-Octadecadienoic acid (Z,Z)- (Retention time 38.205 min), which accounts for 14.44%. The third major ingredient in PSE was 9,12,15-Octadecatrienoic acid (Z,Z,Z)- (Retention time 38.329 min), accounting for 11.97%. Both 9,12-Octadecadienoic acid (Z,Z)- and 9,12,15-Octadecatrienoic acid (Z,Z,Z)- have been reported to reduce the production of NO, and in particular, 9,12-Octadecadienoic acid (Z,Z)- was found to suppress the production of TNF-α, IL-1β, and IL-6 (Lee et al., 2011).
Furthermore, it was established that numerous compounds contained in PSE have anti-inflammatory activities, as Table 2 indicates. Therefore, it can be inferred that the anti-inflammatory activity of PSE demonstrated in the previous experiments may be caused by the presence of these compounds.
This study investigated the anti-inflammatory effects of PSE on LPS-stimulated RAW 264.7 cells. PSE effectively reduced LPS-induced inflammation through the regulation of NF-κB and MAPK signaling pathways. Additionally, PSE inhibited the production of key inflammatory mediators and pro-inflammatory cytokines. Finally, GC-MS analysis was used to determine the chemical composition of PSE, and among the 38 compounds identified, 25 were reported to have anti-inflammatory activity. The results of this study demonstrated the anti-inflammatory activity of P. schrenkii at the molecular level, suggesting the possibility of using P. schrenkii to develop as an anti-inflammatory agent.
Acknowledgments
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea(NRF) funded by the Ministry of Education(NRF-2022R1I1A1A01063293).
References
-
Al-Marzoqi AH, Hadi MY and Hameed IH. (2016). Determination of metabolites products by Cassia angustifolia and evaluate antimicrobial activity. Journal of Pharmacognosy and Phytotherapy. 8:25-48.
[https://doi.org/10.5897/JPP2015.0367]

-
Alonso-Castro AJ, Serrano-Vega R, Perez Gutierrez S, Isiordia-Espinoza MA and Solorio-Alvarado CR. (2022). Myristic acid reduces skin inflammation and nociception. Journal of Food Biochemistry. 46:e14013. https://onlinelibrary.wiley.com/doi/full/10.1111/jfbc.14013, (cited by 2024 February 27).
[https://doi.org/10.1111/jfbc.14013]

-
Ameamsri U, Chaveerach A, Sudmoon R, Tanee T, Peigneur S and Tytgat J. (2021). Oleamide in Ipomoea and Dillenia species and inflammatory activity investigated through ion channel inhibition. Current Pharmaceutical Biotechnology. 22:254-261.
[https://doi.org/10.2174/1389201021666200607185250]

-
Aparna V, Dileep KV, Mandal PK, Karthe P, Sadasivan C and Haridas M. (2012). Anti-inflammatory property of n-hexadecanoic acid: Structural evidence and kinetic assessment. Chemical Biology and Drug Design. 80:434-439.
[https://doi.org/10.1111/j.1747-0285.2012.01418.x]

-
Asanka Sanjeewa K, Fernando I, Kim SY, Kim WS, Ahn Gn, Jee Yh and Jeon YJ. (2019). Ecklonia cava(Laminariales) and Sargassum horneri(Fucales) synergistically inhibit the lipopolysaccharide-induced inflammation via blocking NF-κB and MAPK pathways. Algae. 34:45-56.
[https://doi.org/10.4490/algae.2019.34.2.10]

-
Ben Neriah Y and Karin M. (2011). Inflammation meets cancer, with NF-κB as the matchmaker. Nature Immunology. 12:715-723.
[https://doi.org/10.1038/ni.2060]

-
Cao J, Li Q, Shen X, Yao Y, Li L and Ma H. (2021). Dehydroepiandrosterone attenuates LPS-induced inflammatory responses via activation of Nrf2 in RAW264.7 macrophages. Molecular Immunology. 131:97-111.
[https://doi.org/10.1016/j.molimm.2020.12.023]

-
Chakrabarti M and Mukherjee A. (2021). Investigating the underlying mechanism of cadmium-induced plant adaptive response to genotoxic stress. Ecotoxicology and Environmental Safety. 209:111817. https://www.sciencedirect.com/science/article/pii/S0147651320316535, (cited by 2024 March 31).
[https://doi.org/10.1016/j.ecoenv.2020.111817]

-
Chang MC, Chang HH, Wang TM, Chan CP, Lin BR, Yeung SY, Yeh CY, Cheng RH and Jeng JH. (2014). Antiplatelet effect of catechol is related to inhibition of cyclooxygenase, reactive oxygen species, ERK/p38 signaling and thromboxane A2 production. PLOS ONE. 9:e104310. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0104310, (cited by 2024 February 27).
[https://doi.org/10.1371/journal.pone.0104310]

-
Chen L, Deng H, Cui H, Fang J, Zuo Z, Deng J, Li Y, Wang X and Zhao L. (2018). Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 9:7204-7218.
[https://doi.org/10.18632/oncotarget.23208]

-
Chinnadurai V, Viswanathan P, Kalimuthu K, Vanitha A, Ranjitha V and Pugazhendhi A. (2019). Comparative studies of phytochemical analysis and pharmacological activities of wild and micropropagated plant ethanol extracts of Manihot esculenta. Biocatalysis and Agricultural Biotechnology. 19:101166. https://www.sciencedirect.com/science/article/abs/pii/S1878818119304517, (cited by 2024 February 27).
[https://doi.org/10.1016/j.bcab.2019.101166]

- Chippada SC, Volluri SS, Bammidi SR and Vangalapati M. (2011). In vitro Anti-inflammatory activity of methanolic extract of centella asiatica by HRBC membrane stabilisation. Rasayan Journal of Chemistry. 4:457-460.
-
Chuang LT, Shih YH, Huang WC, Lin LC, Hsu C, Chyuan JH, Tsai TH and Tsai PJ. (2020). In vitro and in vivo screening of wild bitter melon leaf for anti-inflammatory activity against Cutibacterium acnes. Molecules. 25:4277. https://www.mdpi.com/1420-3049/25/18/4277, (cited by 2024 February 27).
[https://doi.org/10.3390/molecules25184277]

-
Closse A, Haefliger W, Hauser D, Gubler HU, Dewald B and Baggiolini M. (1981). 2, 3-Dihydrobenzofuran-2-ones: A new class of highly potent anti-inflammatory agents. Journal of Medicinal Chemistry. 24:1465-1471.
[https://doi.org/10.1021/jm00144a019]

-
da Silva RM, Moreira DKT, Zarricueta ML, Macedo JA, Macedo GA and Gambero A. (2019). The postprandial inflammatory response is attenuated by a dietary structured lipid containing behenic acid. Journal of Functional Foods. 58:350-354.
[https://doi.org/10.1016/j.jff.2019.05.013]

-
Das U. (2020). Arachidonic acid has anti-inflammatory and anti-diabetic actions in vitro and in vivo. Current Developments in Nutrition. 4:747-747.
[https://doi.org/10.1093/cdn/nzaa052_016]

-
Dinarello CA. (2000). Proinflammatory cytokines. Chest. 118:503-508.
[https://doi.org/10.1378/chest.118.2.503]

-
Dong L, Yin L, Zhang Y, Fu X and Lu J. (2017). Anti-inflammatory effects of ononin on lipopolysaccharide-stimulated RAW 264.7 cells. Molecular Immunology. 83:46-51.
[https://doi.org/10.1016/j.molimm.2017.01.007]

-
Funk CD. (2001). Prostaglandins and leukotrienes: Advances in eicosanoid biology. Science. 294:1871-1875.
[https://doi.org/10.1126/science.294.5548.1871]

-
Gabay C. (2006). Interleukin-6 and chronic inflammation. Arthritis Research and Therapy. 8:1-6.
[https://doi.org/10.1186/ar1917]

-
Habib S and Ali A. (2011). Biochemistry of nitric oxide. Indian Journal of Clinical Biochemistry. 26:3-17.
[https://doi.org/10.1007/s12291-011-0108-4]

-
Hamed A, Mantawy E, El-Bakly W, Abdel-Mottaleb Y and Azab S. (2020). Methyl palmitate: The naturally occurring cardioprotective agent. Archives of Pharmaceutical Sciences Ain Shams University. 4:47-62.
[https://doi.org/10.21608/APS.2020.2003.1026]

-
Holanda Pinto S, Pinto L, Cunha G, Chaves M, Santos F and Rao V. (2008). Anti-inflammatory effect of α, β-amyrin, a pentacyclic triterpene from Protium heptaphyllum in rat model of acute periodontitis. Inflammopharmacology. 16:48-52.
[https://doi.org/10.1007/s10787-007-1609-x]

-
Insuan O, Thongchuai B, Khamchun S, Insuan W, Daorueang D and Sansai P. (2022). Antidesma thwaitesianum Müll. Arg. fruit extract rich in 5-hydroxymethylfurfural exhibits anti-inflammatory effects in lipopolysaccharide-stimulated RAW264.7 macrophages. Journal of Herbmed Pharmacology. 11:278-285.
[https://doi.org/10.34172/jhp.2022.33]

-
Islam MT, Ali ES, Uddin SJ, Shaw S, Islam MA, Ahmed MI, Shill MC, Karmakar UK, Yarla NS and Khan IN. (2018). Phytol: A review of biomedical activities. Food and Chemical Toxicology. 121:82-94.
[https://doi.org/10.1016/j.fct.2018.08.032]

- Jananie R, Priya V and Vijayalakshmi K. (2011). Determination of bioactive components of Cynodon dactylon by GC-MS analysis. New York Science Journal. 4:16-20.
- Jayawardena TU, Kim HS, Sanjeewa KA, Han EJ, Jee YH, Ahn GN, Rho JR and Jeon YJ. (2021). Loliolide, isolated from Sargassum horneri; abate LPS-induced inflammation via TLR mediated NF-κB, MAPK pathways in macrophages. Algal Research. 56:102297. https://www.sciencedirect.com/science/article/abs/pii/S2211926421001168, (cited by 2024 February 27).
- Kim AL, Labasi JM, Zhu Y, Tang X, McClure K, Gabel CA, Athar M and Bickers DR. (2005). Role of p38 MAPK in UVB-induced inflammatory responses in the skin of SKH-1 hairless mice. Journal of Investigative Dermatology. 124:1318-1325.
-
Kim BH, Oh I, Kim JH, Jeon JE, Jeon B, Shin J and Kim TY. (2014). Anti-inflammatory activity of compounds isolated from Astragalus sinicus L. in cytokine-induced keratinocytes and skin. Experimental and Molecular Medicine. 46:e87. https://www.nature.com/articles/emm2013157, (cited by 2024 January 10).
[https://doi.org/10.1111/j.0022-202X.2005.23747.x]

-
Kim EA, Kim SY, Kim JS, Oh JY, Kim HS, Yoon WJ, Kang DH and Heo SJ. (2019). Tuberatolide B isolated from Sargassum macrocarpum inhibited LPS-stimulated inflammatory response via MAPKs and NF-κB signaling pathway in RAW264.7 cells and zebrafish model. Journal of Functional Foods. 52:109-115.
[https://doi.org/10.1038/emm.2013.157]

-
Kim EA, Kim SY, Ye BR, Kim Js, Ko SC, Lee WW, Kim KN, Choi IW, Jung WK and Heo SJ. (2018). Anti-inflammatory effect of Apo-9′-fucoxanthinone via inhibition of MAPKs and NF-κB signaling pathway in LPS-stimulated RAW 264.7 macro phages and zebrafish model. International Immunopharmacology. 59:339-346.
[https://doi.org/10.1016/j.jff.2018.10.030]

-
Kim J, Kim JH, Bang SI, Shin H, Cho EJ and Lee S. (2022). Antioxidant activity of edible sprouts and phytosterol contents by HPLC/UV analysis. Horticulture, Environment, and Biotechnology. 63:769-778.
[https://doi.org/10.1016/j.intimp.2018.03.034]

-
Kim J, Nguyen QN, Shin H, Kang KS and Lee S. (2021). In vitro Anti-inflammatory activities and phenolic acid analysis of tree sprout extracts. Korean Journal of Pharmacognosy. 52:257-266.
[https://doi.org/10.1007/s13580-022-00434-6]

- Konishi I, Hosokawa M, Sashima T, Maoka T and Miyashita K. (2008). Suppressive effects of alloxanthin and diatoxanthin from Halocynthia roretzi on LPS-induced expression of pro-inflammatory genes in RAW264.7 cells. Journal of Oleo Science. 57:181-189.
-
Kumar PP, Kumaravel S and Lalitha C. (2010). Screening of antioxidant activity, total phenolics and GC-MS study of Vitex negundo. African Journal of Biochemistry Research. 4:191-195.
[https://doi.org/10.5650/jos.57.181]

- Lee CK, Kim H, Moon KH and Shin KH. (1998). Screening and isolation of antibiotic resistance inhibitors from herb materials-resistance inhibition of volatile components of Korean aromatic herbs. Archives of Pharmacal Research. 21:62-66.
-
Lee SM, Lee TH, Cui EJ, Baek NI, Hong SG, Chung IS and Kim JY. (2011). Anti-inflammatory effects of cowpea(Vigna sinensis K.) seed extracts and its bioactive compounds. Journal of the Korean Society for Applied Biological Chemistry. 54:710-717.
[https://doi.org/10.1007/BF03216754]

-
Liu X, Yin S, Chen Y, Wu Y, Zheng W, Dong H, Bai Y, Qin Y, Li J and Feng S. (2018). LPS-induced proinflammatory cytokine expression in human airway epithelial cells and macrophages via NF-κB, STAT3 or AP-1 activation. Molecular Medicine Reports. 17:5484-5491.
[https://doi.org/10.1007/BF03253149]

-
Marasinghe CK, Jung WK and Je JY. (2022). Anti-inflammatory action of ark shell(Scapharca subcrenata) protein hydrolysate in LPS-stimulated RAW264.7 murine macrophages. Journal of Food Biochemistry. 46:e14493. https://onlinelibrary.wiley.com/doi/full/10.1111/jfbc.14493, (cited by 2024 January 15).
[https://doi.org/10.3892/mmr.2018.8542]

-
Mazimba O. (2017). Umbelliferone: Sources, chemistry and bioactivities review. Bulletin of Faculty of Pharmacy, Cairo University. 55:223-232.
[https://doi.org/10.1111/jfbc.14493]

-
Miyasaka N and Hirata Y. (1997). Nitric oxide and inflammatory arthritides. Life Sciences. 61:2073-2081.
[https://doi.org/10.1016/j.bfopcu.2017.05.001]

-
Mohammed GJ, Omran AM and Hussein HM. (2016). Antibacterial and phytochemical analysis of Piper nigrum using gas chromatography-mass spectrum and Fourier-transform infrared spectroscopy. International Journal of Pharmacognosy and Phytochemical Research. 8:977-996.
[https://doi.org/10.1016/S0024-3205(97)00585-7]

- Munn LL. (2017). Cancer and inflammation. WIREs Mechanisms of Disease 9:e1370. https://wires.onlinelibrary.wiley.com/doi/abs/10.1002/wsbm.1370, (cited by 2024 March 31).
-
Myung DB, Lee JH, Han HS, Lee KY, Ahn HS, Shin YK, Song EJ, Kim BH, Lee KH and Lee SH. (2020). Oral intake of hydrangea serrata(Thunb.) ser. leaves extract improves wrinkles, hydration, elasticity, texture, and roughness in human skin: A randomized, double-blind, placebo-controlled study. Nutrients. 12:1588. https://www.mdpi.com/2072-6643/12/6/1588, (cited by 2024 April 11).
[https://doi.org/10.1002/wsbm.1370]

-
Nakamura S, Matsuda H and Yoshikawa M. (2011). Search for antidiabetic constituents of medicinal food. Journal of the Pharmaceutical Society of Japan. 131:909-915.
[https://doi.org/10.3390/nu12061588]

-
Newman DJ and Cragg GM. (2020). Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. Journal of Natural Products. 83:770-803.
[https://doi.org/10.1248/yakushi.131.909]

-
Newton K and Dixit VM. (2012). Signaling in innate immunity and inflammation. Cold Spring Harbor Perspectives in Biology. 4:a006049. https://cshperspectives.cshlp.org/content/4/3/a006049.short, (cited by 2024 February 2).
[https://doi.org/10.1021/acs.jnatprod.9b01285]

-
Nore KG, Jørgensen MJ, Dyrhol-Riise AM, Jenum S and Tonby K. (2020). Elevated levels of anti-inflammatory eicosanoids and monocyte heterogeneity in Mycobacterium tuberculosis infection and disease. Frontiers in immunology. 11:579849. https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2020.579849/full, (cited by 2024 March 31).
[https://doi.org/10.1101/cshperspect.a006049]

-
Nunes CDR, Barreto Arantes M, Menezes de Faria Pereira S, Leandro da Cruz L, de Souza Passos M, Pereira de Moraes L, Vieira IJC and Barros de Oliveira D. (2020). Plants as sources of anti-inflammatory agents. Molecules. 25:3726. https://www.mdpi.com/1420-3049/25/16/3726, (cited by 2024 Febrary 2).
[https://doi.org/10.3389/fimmu.2020.579849]

-
Okur ME, Karadağ AE, Üstündağ Okur N, Özhan Y, Sipahi H, Ayla Ş, Daylan B, Demirci B and Demirci F. (2020). In vivo wound healing and in vitro anti-inflammatory activity evaluation of Phlomis russeliana extract gel formulations. Molecules. 25:2695. https://www.mdpi.com/1420-3049/25/11/2695, (cited by 2024 February 27).
[https://doi.org/10.3390/molecules25163726]

-
Olukanni A. (2020). Antioxidant and in vitro anti-inflammatory activities of Albizia zygia(DC) JF mebr and the evaluation of its phytochemical constituents. Journal of Medicinal Plants Studies. 8:317-323.
[https://doi.org/10.3390/molecules25112695]

- Omeje K, Ozioko J and Opmeje H. (2018). Pharmacological potentials, characterization and fatty acids profile of Persea americana Mill.(avocardo) seed oil using gas chromatography-mass spectroscopy. Biochemistry and Analytical Biochemistry. 7:1000361. https://www.researchgate.net/profile/Kingsley-Omeje/publication/331225970_Pharmacological_Potentials_Charac-terization_and_Fatty_Acids_Profile_of_Persea_americana_Mill_Avocardo_Seed_Oil_Using_Gas_Chromatography-Mass_Spectroscopy/links/5f2a87d792851cd302dc62e5/Pharmacological-Potentials-Characterization-and-Fatty-Acids-Profile-of-Persea-americana-Mill-Avocardo-Seed-Oil-Using-Gas-Chromatography-Mass-Spectroscopy.pdf, (cited by 2024 February 27).
- Park S, Kim H, Lee HS and Chang CS. (2005). Taxonomic reconsideration of the Philadelphus schrenkii complex. Korean Journal of Plant Taxonomy. 35:247-272.
-
Rajeswari G, Murugan M and Mohan V. (2012). GC-MS analysis of bioactive components of Hugonia mystax L.(Linaceae). Research Journal of Pharmaceutical, Biological and Chemical Sciences. 3:301-308.
[https://doi.org/10.11110/kjpt.2005.35.4.247]

- Ramadass V, Vaiyapuri T and Tergaonkar V. (2020). Small molecule NF-κB pathway inhibitors in clinic. International Journal of Molecular Sciences. 21:5164. https://www.mdpi.com/1422-0067/21/14/5164, (cited by 2024 April 11).
-
Seo J, Lee U, Seo S, Wibowo AE, Pongtuluran OB, Lee K, Han SB and Cho S. (2022). Anti-inflammatory and antioxidant activities of methanol extract of Piper betle Linn.(Piper betle L.) leaves and stems by inhibiting NF-κB/MAPK/Nrf2 signaling pathways in RAW 264.7 macrophages. Biomedicine and Pharmacotherapy. 155:113734. https://www.sciencedirect.com/science/article/pii/S0753332222011234, (cited by 2024 Fabruary 12).
[https://doi.org/10.3390/ijms21145164]

-
Shalini K and Ilango K. (2021). Preliminary phytochemical studies, GC-MS analysis and in vitro antioxidant activity of selected medicinal plants and its polyherbal formulation. Pharmacognosy Journal. 13:648-659.
[https://doi.org/10.1016/j.biopha.2022.113734]

-
Sharma JN, Al-Omran A and Parvathy SS. (2007). Role of nitric oxide in inflammatory diseases. Inflammopharmacology. 15:252-259.
[https://doi.org/10.5530/pj.2021.13.83]

-
Shi J, Li CJ, Yang JZ, Ma J, Li Y, Chen H and Zhang DM. (2015). Monoterpenes from the leaves of Hydrangea paniculata and their hepatoprotective activities. Journal of Asian Natural Products Research. 17:512-518.
[https://doi.org/10.1007/s10787-007-0013-x]

-
Shin KH, Chi HJ, Lim SS, Cho SH, Moon HI and Yu JH. (1997). Antimicrobial activities of volatile essential oils from Korean aromatic plants. Natural Product Sciences. 3:141-147.
[https://doi.org/10.1080/10286020.2015.1042871]

- Simon LS. (1999). Role and regulation of cyclooxygenase-2 during inflammation. The American Journal of Medicine. 106:37S-42S.
-
Smale ST. (2011). Hierarchies of NF-κB target-gene regulation. Nature Immunology. 12:689-694.
[https://doi.org/10.1016/S0002-9343(99)00115-1]

-
Sung YY, Kim MS, Yuk HJ, Kim SH, Lee GJ and Kim DS. (2023). Anti-obesity effects of Philadelphus tenuifolius extract via improvement of lipid metabolism in high-fat diet-induced obese mice. Planta Medica. 89:P-273. https://www.thieme-connect.com/products/ejournals/html/10.1055/s-0043-1774144, (cited by 2024 April 11).
[https://doi.org/10.1038/ni.2070]

- Taciak B, Białasek M, Braniewska A, Sas Z, Sawicka P, Kiraga Ł, Rygiel T and Król M. (2018). Evaluation of phenotypic and functional stability of RAW 264.7 cell line through serial passages. PLOS ONE. 13:e0198943. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0198943, (cited by 2024 February 12).
-
Tsai FS, Lin LW and Wu CR. (2016). Lupeol and its role in chronic diseases. Drug Discovery from Mother Nature. 2016:145-175.
[https://doi.org/10.1371/journal.pone.0198943]

-
Vadivel E and Gopalakrishnan S. (2011). GC-MS analysis of some bioactive constituents of Mussaenda frondosa Linn. International Journal of Pharma and Bio Sciences. 2:313-320.
[https://doi.org/10.1007/978-3-319-41342-6_7]

- Venn-Watson SK and Butterworth CN. (2022). Broader and safer clinically-relevant activities of pentadecanoic acid compared to omega-3: Evaluation of an emerging essential fatty acid across twelve primary human cell-based disease systems. PLOS ONE. 17:e0268778. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0268778, (cited by 2024 February 27).
-
Viatour P, Merville MP, Bours V and Chariot A. (2005). Phosphorylation of NF-κB and IκB proteins: Implications in cancer and inflammation. Trends in Biochemical Sciences. 30:43-52.
[https://doi.org/10.1371/journal.pone.0268778]

-
Vuolteenaho K, Moilanen T, Knowles RG and Moilanen E. (2007). The role of nitric oxide in osteoarthritis. Scandinavian Journal of Rheumatology. 36:247-258.
[https://doi.org/10.1016/j.tibs.2004.11.009]

-
Wang J, Liu Y, Gu Y, Liu C, Yang Y, Fan X, Yang H, Liu Y and Ma T. (2023). Function and inhibition of P38 MAP kinase signaling: Targeting multiple inflammation diseases. Biochemical
[https://doi.org/10.1080/03009740701483014]

-
Wink DA, Hines HB, Cheng RY, Switzer CH, Flores-Santana W, Vitek MP, Ridnour LA and Colton CA. (2011). Nitric oxide and redox mechanisms in the immune response. Journal of Leukocyte Biology. 89:873-891.
[https://doi.org/10.1016/j.bcp.2023.115973]

-
Xiao P. (1989). Zhongguo ben cao tu lu(Vol. 7). Ren min wei sheng chu ban she. Beijing, China. p.68.
[https://doi.org/10.1189/jlb.1010550]

- Yadav A, Saini V and Arora S. (2010). MCP-1: Chemoattractant with a role beyond immunity: A review. Clinica Chimica Acta. 411:1570-1579.
-
Yang GS, Lee KJ, Lee MH, Ham IH and Choi HY. (2012). Inhibition of lipopolysaccharide-induced nitric oxide and prostaglandin E2 production by chloroform fraction of Cudrania tricuspidata in RAW 264.7 macrophages. BMC Complementary and Alternative Medicine. 12:1-7.
[https://doi.org/10.1016/j.cca.2010.07.006]

-
Zhang D, Guo H, Feng W and Qiu H. (2019). LAMC2 regulated by microRNA-125a-5p accelerates the progression of ovarian cancer via activating p38 MAPK signalling. Life Sciences. 232:116648. https://www.sciencedirect.com/science/article/abs/pii/S0024320519305740, (cited by 2024 February 15).
[https://doi.org/10.1186/1472-6882-12-250]