
Development of Insertions and deletions (InDel) Markers for the Differentiation of Cnidium officinale Makino and Ligusticum chuanxiong Hort
This is an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
In Korea, the medicinal herb Cnidii Rhizoma is manufactured using Cnidium officinale Makino and Ligusticum chuanxiong Hort., with the former being the primary ingredient. C. officinale and L. chuanxiong have similar external appearances, and because their roots are dried for use as medicinal materials, it is difficult to distinguish between them. Therefore, there is a need to develop easy-to-use molecular markers to facilitate the classification of Cnidii Rhizoma varieties and the identification of medicinal materials.
DNA was extracted from C. officinale and L. chuanxiong and analyzed using an Illumina HiSeq 2500 platform. After de novo assembly, the loci showing polymorphisms between C. officinale and L. chuanxiong were identified. Primers were designed for polymorphic loci, and genotyping was performed. Finally, three nuclear genome-based insertion and deletion (InDel) markers, CoInDel-01, CoInDel-02, and CoInDel-03, were developed. These markers produced a single band at 230, 369, and 245 bp, respectively, in C. officinale, whereas two bands were observed in L. chuanxiong at 230 and 264 bp, 304 and 369 bp, and 197 and 245 bp, respectively.
The three InDel markers developed in this study will be useful for differentiating between C. officinale and L. chuanxiong.
Keywords:
Cnidium officinale, Genotyping, Insertion and Deletion, Ligusticum chuanxiong, Next Generation SequencingINTRODUCTION
Cnidium officinale Makino (Synonyms: Ligusticum officinale (Makino) Kitagawa, Conioselinum officinale (Makino) Ohashi & Ohashi) (Kitagawa, 1963; Ohashi and Ohashi, 2023), a perennial herb of Apiaceae, occurs in Korea, and has been widely cultivated in East Asia for a long time as medicinal plant. It was written as ‘川芎’ in Chinese, the old East Asia character, and is called ‘Cheongung’ in Korean and ‘Senkyu’ in Japanese. In China, ‘川芎’ is called Chuanxiong, which refers to Ligusticum chuanxiong (Yoon et al., 2023; Song et al., 2024).
According to the Korean Pharmacopoeia (MFDS, 2022), Cnidii Rhizoma is primarily made from the root stems of C. officinale or L. chuanxiong Hort. ex Qiu, et al.
Cnidii Rhizoma have been used in the treatment of headaches, anemia, pain, and gynecological disorders, and their medicinal effects are being re-evaluated in modern medicine (Jeong et al., 2015). In particular, research on its antioxidant, anti-inflammatory, and antimicrobial effects is actively ongoing (Jeong et al., 2009; Um et al., 2017; Jeong et al., 2020; Lee et al., 2021). The major bioactive compounds found in Cnidii Rhizoma include cnidilide, ligustilide, and senkyunolide (Song et al., 2009).
Cnidii Rhizoma is referred to as C. officinale or L. chuanxiong, depending on the source plant (Song et al., 2009; Kim et al., 2020a). While C. officinale and L. chuanxiong are known to belong to the same lineage, there is controversy regarding the origin plant due to the indistinct differences between C. officinale and the others (Park, 1998; Song et al., 2009). C. officinale and L. chuanxiong have similar plant appearances, making it difficult to distinguish them by sight (Lee et al., 1999; Jung et al., 2019).
C. officinale cultivated in Korea, its stems are softer, with several small tubers fused together to form a circular shape, in contrast to L. chuanxiong (Kim et al., 2020b). L. chuanxiong is cultivated and distributed in Korea, but its botanical classification remains unclear, as it belongs to the genus Angelica (Suh et al., 2016; Kim et al., 2020b). Morphologically, it has rhizomes that develop into thin, fibrous root-like structures and grows larger than C. officinale (Jung et al., 2019; Kim et al., 2020a).
These two species have similar appearances, making accurate identification necessary. Furthermore, with the increasing demand for medicinal herbs, imports have risen, leading to confusion in the domestic distribution process. In the distribution process, issues have arisen due to unclear origins or similar names of medicinal plants (Han et al., 2015).
To address these issues, accurate identification, effective differentiation, and quality control procedures for medicinal plants are required. Recently, with the development of next generation sequencing (NGS) technology, Illumina, PacBio, Nanopore, etc., large amounts of data can be generated quickly and analyzed at a low cost. This enables the comparison of genetic information across different species, allowing for the study of the evolutionary relationships of plants (Han et al., 2015).
DNA molecular markers are used to identify or differentiate organisms at the molecular level based on their nucleotide sequences (Huh, 2015). DNA molecular markers include SNP (single nucleotide polymorphism), InDel (insertion and deletion), SSR (simple sequence repeat), RAPD (random amplified polymorphic DNA), RFLP (restriction fragment length polymorphism), among others (Al-Samarai et al, 2015). The appropriate marker is selected depending on the purpose of the research.
To date, molecular markers developed for Cnidii Rhizoma include studies on RAPD and RFLP markers, and an SNP marker has been developed in the internal transcribed spacer (ITS) region of the nuclear ribosomal RNA gene (Ji et al, 2017). However, there are no reported cases of InDel marker development (Song et al., 2009; Kim et al., 2014). InDel markers can be easily detected through PCR (polymerase chain reaction), offering the advantage of simple analysis by comparing the size of the amplified products using gel electrophoresis (Kim et al., 2022; Kim et al., 2023).
This study aimed to clearly differentiate C. officinale and L. chuanxiong by developing InDel markers based on the sequences obtained using Illumina HiSeq 2500 platform from the plants cultivated in Korea.
Materials and Methods
1. Materials
In this experiment, young leaves from a total of 11 individuals, including 8 C. officinale and 3 L. chuanxiong cultivated in Korea, were collected and used (Table 1 and Fig. 1).
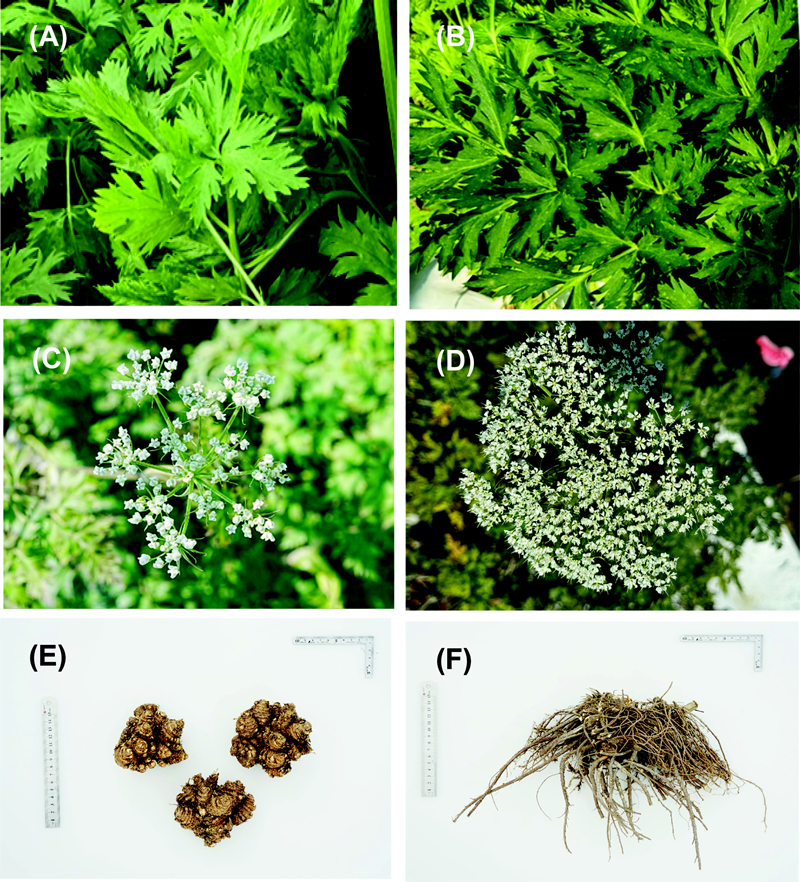
Photograph of C. officinale (A, C, E) and L. chuanxiong (B, D, F).(A, B); Young leaves, (C, D); Flowers, (E, F); Rhizome.
The leaves of C. officinale are lighter green and the mesophyll is thinner (Fig. 1A) than that of L. chuanxiong (Fig. 1B). The flower of C. officinale is a compound umbel and consists of about 10 to 13 inflorescences, with about 10 to 15 flowers per inflorescence (Fig. 1C), whereas the flower of L. chuanxiong is a compound umbel and consists of about 20 inflorescences, with about 25 to 30 flowers per inflorescence (Fig. 1D). The rhizome of C. officinale enlarges and forms a bulb (Fig. 1E), but that of L. chuanxiong develops thin and long (Fig. 1F).
2. DNA extraction
DNA extraction from the 11 individuals of C. officinale and L. chuanxiong was performed using the BiomedicⓇ Plant gDNA Extraction Kit (Biomedic Co., Ltd., Bucheon, Korea) according to the manufacturer’s instructions. DNA concentration was measured using NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and adjusted to 10 ng/㎕ before storage at –20℃.
3. Next-generation sequencing (NGS) analysis
Next-generation sequencing (NGS) analysis was performed on one individual of C. officinale (Co2401_1) and one individual of L. chuanxiong (Co2415_1), with paired-end libraries constructed using the TruSeq Nano DNA Sample Prep Kit according to the manufacturer's manual (Illumina, Inc., San Diego, CA, USA). Paired-end sequencing with a 151-bp read length conducted using the Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA).
The raw data obtained from the NGS results were processed to improve quality, and the preprocessed reads were assembled de novo using the CLC Genomics Workbench ver. 20.0 (Qiagen, Aarhus, Denmark) software to obtain contigs.
4. InDel region identification and primer design
The contigs over than 10 kbp were selected from the assembled contigs of C. officinale were compared to the corresponding contigs of L. chuanxiong using alignment function of the CLC Genomics Workbench ver. 20.0 software to identify polymorphic InDel regions, and primers were designed for these regions using the same software.
The primer design conditions were as follows: length of 18 bp to 22 bp, annealing temperature of 48℃ to 60℃, GC content between 30% and 60%, and product size ranging from 150 bp to 450 bp.
5. Polymerase chain reaction (PCR) analysis
The composition of the PCR reaction mixture consisted of 1 ㎕ of gDNA, 1 ㎕ each of the forward and reverse primers, 10 ㎕ of 2 × Taq mix (Dongsheng Biotech Co, Ltd, Guangzhou, Guangdong, China) and 7 ㎕ of distilled water, for a total volume of 20 ㎕.
Amplification was carried out using a T100™ Thermal Cycler (Bio-Rad Inc., Hercules, CA, USA). The PCR conditions were as follows: pre-denaturation at 95℃ for 3 minutes, followed by 35 cycles of denaturation at 95℃ for 30 seconds, annealing at 54℃ or 55℃ for 30 seconds, and extension at 72℃ for 1 minute. A final extension was performed at 72℃ for 5 minutes. The PCR products were stored at –20℃.
6. Electrophoresis
Electrophoresis was performed to determine the genotypes of the PCR amplification products. The agarose gel used was prepared with 1 × TAE buffer and agarose at a concentration of 3%. DNA visualization was achieved using EtBr (ethidium bromide).
As a size marker, the 1 kb Ladder Plus (Dongsheongbio Company Ltd., Guangdong, China) was used to determine the size of the amplified products.
Electrophoresis was conducted for 30 minutes at 120 V using the Mini-sub Cell GT System (Bio-Rad Inc., Hercules, CA, USA), and the bands were visualized using the Gel Doc™ XR+ System (Bio-Rad Inc., Hercules, CA, USA).
7. Mapping of NGS data to the amplicons of three markers
The NGS data produced from L. chuanxiong were trimmed using the CLC Genomics Workbench ver. 20.0 (Qiagen, Aarhus, Denmark) software and then mapped to the nucleotide sequences amplified by each marker.
Mapping was performed using the following parameters (Match score = 1; Mismatch cost = 2; Cost of insertions and deletions = Linear gap cost; Insertion cost = 3; Deletion cost = 3; Length fraction = 0.8; Similarity fraction = 0.9; Non-specific match handling = Map randomly; Minimum seed length = 15).
RESULTS
1. NGS analysis and InDel region exploration results
According to the NGS analysis, the total read size of C. officinale (Co2401_1) was 68,358,861,794 bp, with a total of 452,707,694 reads, and the GC content was 36.4%. The total read size of L. chuanxiong (Co2415_1) was 51,704,152,808 bp, with a total of 342,411,608 reads, and the GC content was 36.8%.
A comparative analysis of the DNA sequences of C. officinale and L. chuanxiong was conducted to explore the regions exhibiting polymorphism. A total of 20 candidate InDel loci were selected, and primers were designed.
PCR analysis was performed to observe whether the designed primer sets amplified correctly and to assess the genotypes, and polymorphism was analyzed through gel electrophoresis. The electrophoresis genotyping results revealed the development of three InDel markers capable of distinguishing between C. officinale and L. chuanxiong, which were named CoInDel-01, CoInDel-02, and CoInDel-03 (Table 2).
2. Genotyping using developed InDel markers
Genotyping of a total of 11 individuals, including 8 C. officinale (Co2401-1, Co2402-1, Co2403-1, Co2405-1, Co2410-1, Co2411-1, Co2413-1, Co2414-1) and 3 L. chuanxiong (Co2415-1, Co2415-2, Co2415-3), was performed using the CoInDel-01 marker. The results showed a 230 bp band in all C. officinale individuals, while the L. chuanxiong individuals exhibited both a 230 bp band and an additional 269 bp band (Fig. 2A).
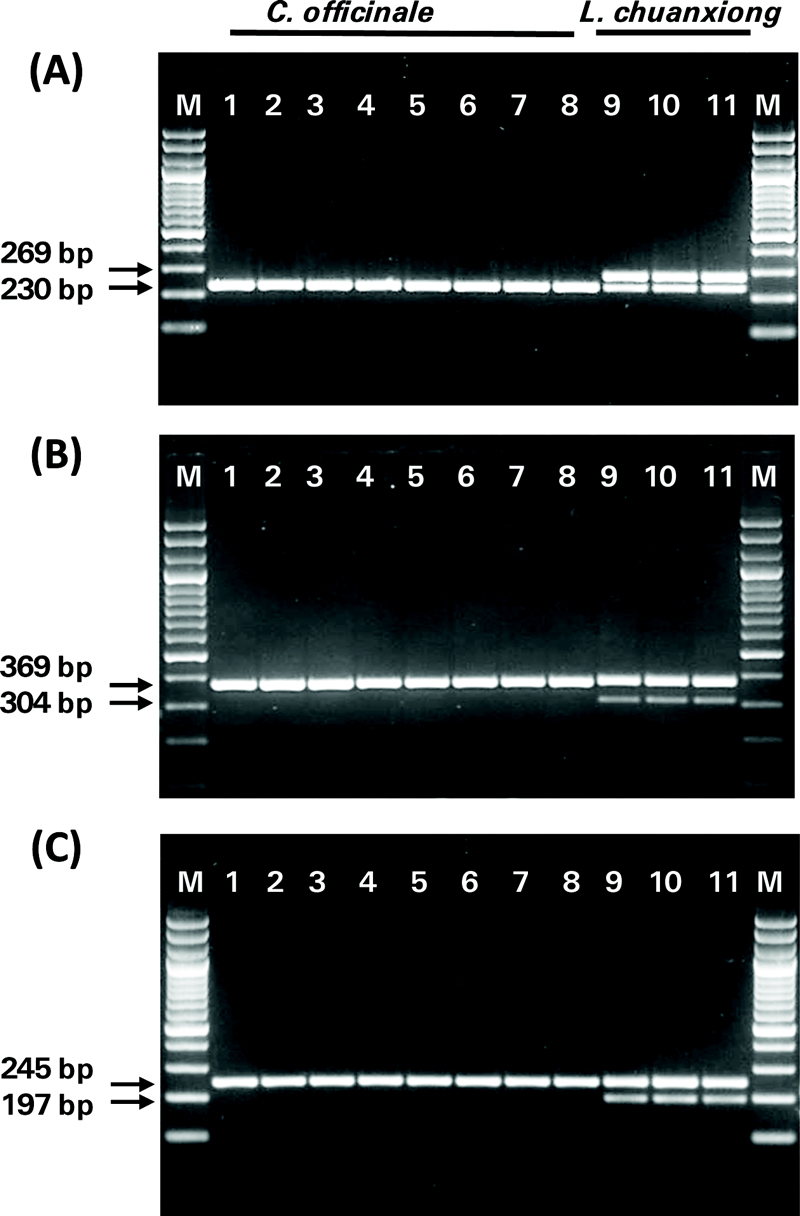
Results of electrophoresis performed on 11 genetic resources using 3 InDel markers for the distinguishment of C. officinale and L. chuanxiong. (A); CoInDel-01, (B); CoInDel-02, and (C); CoInDel-03, Lane M; size marker GB-Ruler™ 100 bp Plus DNA Ladder (GeneBio Systems, Montreal, QC, Canada), Lane 1; Co2401-1, Lane 2; Co2402-1, Lane 3; Co2403-1, Lane 4; Co2405-1, Lane 5; Co2410-1. Lane 6; Co2411-1, Lane 7; Co2413-1, Lane 8; Co2414-1, Lane 9; Co2415-1, Lane 10; Co2415-2, Lane 11; Co2415-3.
Genotyping using the CoInDel-02 marker showed that a single band appeared at 369 bp in all C. officinale individuals (Co2401-1, Co2402-1, Co2403-1, Co2405-1, Co2410-1, Co2411-1, Co2413-1, Co2414-1), while in the L. chuanxiong individuals (Co2415-1, Co2415-2, Co2415-3), an additional band was observed at 304 bp along with the 369 bp band (Fig. 2B).
Genotyping using the CoInDel-03 marker showed that a single band was amplified at 245 bp in all C. officinale individuals (Co2401-1, Co2402-1, Co2403-1, Co2405-1, Co2410-1, Co2411-1, Co2413-1, Co2414-1), while in the L. chuanxiong individuals (Co2415-1, Co2415-2, Co2415-3), two bands were amplified at 245 bp and 197 bp (Fig. 2C).
In all InDel markers, a single band (230 bp, 369 bp, 245 bp) was observed in C. officinale, while two bands (269 bp/230 bp, 369 bp/304 bp, 245 bp/197 bp) were observed in L. chuanxiong. Each analysis using the three InDel markers clearly distinguished between C. officinale and L. chuanxiong.
DISCUSSION
Existing markers have been used to distinguish between C. officinale and L. chuanxiong, but there are limitations with these markers. For example, RAPD and RFLP markers suffer from reproducibility issues, and SNP markers developed in the ITS region have limited application issue (Ji et al, 2017). Therefore, the development of additional DNA molecular markers was necessary.
In this study, we developed InDel markers that can easily distinguish between C. officinale and L. chuanxiong. InDel markers can effectively differentiate plant species (Garcia-Lor et al. 2013; Yamaki et al. 2013; Cho et al. 2015; Kim et al. 2021). Examples of species or cultivar identification using InDel markers include the differentiation of Schisandra (Jeong et al., 2021), differentiation of domestic jujube varieties (Kim et al., 2021), distinction between Prunus mume, Prunus armeniaca var. ansu, and their interspecific hybrids (Kim et al., 2022), differentiation between Paeonia (Lee et al., 2022), identification of Codonopsis species (Kim et al., 2023), and identification of plants in the genus Broussonetia (Lee et al., 2023).
In this study, 8 C. officinale individuals and 3 L. chuanxiong individuals were analyzed using InDel markers through PCR and gel electrophoresis. The results showed that all three markers clearly distinguished between C. officinale and L. chuanxiong.
Through additional analysis, the Illumina sequencing data of L. chuanxiong (Co2415-1) produced in this study were mapped to the sequences amplified by each marker. The ratio of the coverage of common amplicons to the coverage of L. chuanxiong-specific amplicons was calculated for the three markers developed in this study. The ratios for CoInDel-01, CoInDel-02, and CoInDel-03 were 1.80, 2.14, and 2.39, respectively (Table 3). This ratio can be seen as the coverage of the amplicon commonly found in C. officinale and L. chuanxiong being about twice the coverage of the amplicon appearing only in L. chuanxiong.

Read number and coverage of L. chuanxiong NGS data mapped to the amplicon sequences amplified by the markers.
This suggests that the observed 2:1 ratio could be due to L. chuanxiong being a triploid with two chromosomes sharing the same sequence as C. officinale and one chromosome with a different sequence. A chi-square test for goodness of fit, based on the number of reads mapped to each locus, confirmed that the read distribution for all three markers followed a 2 : 1 ratio.
These results in line with previous report that C. officinale is a diploid plant (2n = 22) and L. chuanxiong is a triploid plant (2n = 33) (Song et al., 2021; Suh et al., 2016) and the assumption that L. chuanxiong may be a hybrid that includes the genome of C. officinale (Kondo et al., 1996).
Through the CoInDel-01, CoInDel-02, and CoInDel-03 markers developed in this study, it is now possible to genetically distinguish between C. officinale and L. chuanxiong, which were previously difficult to differentiate.
This will help prevent confusion in the market and provide a foundation for molecular marker-based research in origin studies. Furthermore, it is expected that these markers can be used in the development of superior varieties of C. officinale and L. chuanxiong.
Acknowledgments
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (PJ1732201)” Rural Development Administration, Korea and 2024 year the RDA Fellowship Program National Institute of Horticultural and Herbal Science, Rural Development Administration, Republic of Korea. This research was supported by “Regional Innovation Strategy(RIS)” through the National Research Foundation of Korea(NRF) funded by the Ministry of Education(MOE)(2021RIS-001).
References
-
Al-Samarai FR, Al-Kazaz A.A. (2015). Molecular markers: An introduction and applications. European Journal of Molecular Biotechnology. 9:118-130.
[https://doi.org/10.13187/ejmb.2015.9.118]

-
Cho KS, Yun BK, Yoon YH, Hong SY, Mekapogu M, Kim KH and Yang TJ. (2015). Complete chloroplast genome sequence of tartary buckwheat(Fagopyrum tataricum) and comparative analysis with common buckwheat(F. esculentum). PloS One. 10:e0125332. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.01253321, (cited by 2024 Nov. 21).
[https://doi.org/10.1371/journal.pone.0125332]

-
Garcia-Lor A, Curk F, Snoussi-Trifa H, Morillon R, Ancillo G, Luro F, Navarro L and Ollitrault P. (2013). A nuclear phylogenetic analysis: SNPs, indels and SSRs deliver new insights into the relationships in the ‘true citrus fruit trees’ group(Citrinae, Rutaceae) and the origin of cultivated species. Annals of Botany. 111:1-19.
[https://doi.org/10.1093/aob/mcs227]

-
Han EH, Kim YH and Lee SW. (2015). Development of molecular biological techniques for the differentiation of medicinal plant species. Journal of Plant Biotechnology. 42:6-12.
[https://doi.org/10.5010/JPB.2015.42.1.6]

-
Huh MK. (2015). An overview for molecular markers in plants. Journal of Life Science. 25:839-848.
[https://doi.org/10.5352/JLS.2015.25.7.839]

-
Jeong HJ, Lee JB, Gil JS, Hong CP, Kang SH, Kwon SJ, Kim HJ, Kim CK, Lee JH and Lee Y. (2021). Development of chloroplast-based InDel markers for the discrimination of Schisandraceae plant species. Korean Journal of Medicinal Crop Science. 29:11-16.
[https://doi.org/10.7783/KJMCS.2021.29.1.11]

-
Jeong JB, Ju SY, Park JH, Lee JR, Yun KW, Kwon ST, Lim JH, Chung GY and Jeong HJ. (2009). Antioxidant activity in essential oils of Cnidium officinale Makino and Ligusticum chuanxiong Hort. and their inhibitory effects on DNA damage and apoptosis induced by ultraviolet B in mammalian cell. Cancer Epidemiology. 33:41-46.
[https://doi.org/10.1016/j.canep.2009.04.010]

-
Jeong JT, Ha BK, Han JW, Lee JH, Lee SH, Oh MW, Park CG, Ma KH, Chang JK, Kim SH, Kim JB, Kang SY and Ryu JH. (2020). Gamma-ray irradiation on radio sensitivity in Cnidium officinale Makino. Korean Journal of Medicinal Crop Science. 28:339-346.
[https://doi.org/10.7783/KJMCS.2020.28.5.339]

-
Jeong JW, Choi YH and Park C. (2015). Induction of apoptosis by ethanol extract of Cnidium officinale in human leukemia U937 cells through activation of AMPK. Journal of Life Science. 11:1255-1264.
[https://doi.org/10.5352/JLS.2015.25.11.1255]

-
Ji L, Liu C, Zhang L, Liu A, Yu J. (2017) Variation of rDNA internal transcribed spacer sequences in Rhizoctonia cerealis. Current Microbioloy. 74:877-884.
[https://doi.org/10.1007/s00284-017-1258-2]

-
Jung CR, Jeong DH, Park HW, Kim HJ, Jeon KS and Yoon JB. (2019). Molecular identification of thrips in two medicinal crops, Cnidium officinale Makino and Ligusticum chuanxiong Hort. Korean Journal of Medicinal Crop Science. 27:17-23.
[https://doi.org/10.7783/KJMCS.2019.27.1.17]

-
Kim JH, Jeong YH, Lee MS, Kim MK, Jo NS, Gil JS, Koo SC and Lee Y. (2023). Development of InDel Markers to distinguish among Codonopsis species. Korean Journal of Medicinal Crop Science. 31:12-17.
[https://doi.org/10.7783/KJMCS.2023.31.1.12]

-
Kim JH, Lee DW, Kim MK, Lee MS, Jo NS and Lee Y. (2022). Development of InDel markers to distinguish between Prunus mume, Prunus armeniaca var. ansu, and their interspecific hybrids. Korean Journal of Medicinal Crop Science. 30:210-216.
[https://doi.org/10.7783/KJMCS.2022.30.3.210]

-
Kim K, Han KM, Kim HJ, Jeon KS, Kim CW and Jung CR. (2020a). The study of soil chemical properties and soil bacterial communities on the cultivation systems of Cnidium officinale Makino. Korean Journal of Environmental Agriculture 39:1-9.
[https://doi.org/10.5338/KJEA.2020.39.1.1]

-
Kim MK, Kim JH, Lee MS, Jo NS, Park SI, Gil JS, Yeruult E, Oh HK, Lee KH, Kim HB, Lee MS and Lee Y. (2021). Development of insertion or deletion markers to distinguish Korean jujube cultivars. Korean Journal of Medicinal Crop Science. 29:282-292.
[https://doi.org/10.7783/KJMCS.2021.29.4.282]

- Kim NS, Jeon KS and Lee HS. (2020b). Effects of forest environments on growth and active compound contents of Ligusticum chuanxiong Hort. among different forest sites. Korean Journal of Plant Resources. 33:419-427.
-
Kim WJ, Ji Y, Lee YM, Kang YM, Choi G, Kim HK and Moon BC. (2014). Development of molecular marker for the authentication of Patriniae Radix by the analysis of DNA barcodes. Korean Journal of Herbology 29:45-53.
[https://doi.org/10.6116/kjh.2014.29.6.45.]

- Kitagawa M. (1963). Notulae fractae ob floram Asiae Orientalis. The Journal of Japanese Botany. 38:105-111.
-
Kondo K, Terabayashi S, Okada M, Yuan C and He S. (1996). Phylogenetic relationship of medicinally important Cnidium officinale and Japanese Apiaceae based on rbcL sequences. Journal of Plant Research. 109:21-27.
[https://doi.org/10.1007/BF02344283]

- Lee EJ, Kim YA, Lee MS, Kim JH, Choi YK, Kim JS, Shin CS and LEE Y. (2023). Development of chloroplast genome-based insertion/deletion markers in the genus Broussonetia. Korean Journal of Plant Resources. 36:290-298.
- Lee HW, Cho HG and Park YK. (1999) The study on antioxidative effects and quality comparison of Ligusticum chuanziong and Cnidium officinale(2). Korean Journal of Herbology. 14:55-60.
- Lee JW, Kim YS and Lee DS. (2021). A study of new herbal medicine prescription for the treatment of dermatological diseases. Food Industry and Nutrition. 26:36-43.
-
Lee MS, Jeong HJ, Park SH, Chung H, Jeong JT, Kim MK, Gil JS, Lee JB, Kim SR, Yun KH and Lee Y. (2022). Development of a chloroplast-based InDel marker that discriminates between Paeonia suffruticosa and P. lactiflora. Korean Journal of Medicinal Crop Science. 30:430-439.
[https://doi.org/10.7783/KJMCS.2022.30.6.430]

- Ministry of Food and Drug Safety (MFDS). (2022). The Korean P. harmacopoeia. Ministry of Food and Drug Safety. Cheongju, Korea. p.107-108.
- Ohashi K and Ohashi H. (2023). Transfer of Cnidium officinale to Conioselinum(Umbelliferae/Apiaceae). The Journal of Japanese Botany. 98:29-36.
- Park YK. (1998). The study on antioxidative effects and quality comparison of Ligusticum chuanxiong and Cnidium officinale(1). Korean Journal of Herbology. 13:103-114.
-
Song HE, Oh SM, Kim IS, Kim DY, Kim HG, Yoon D, Ryn HW and Lee DY. (2024). Selection of chemical marker for Cnidii Rhizoma by Gyeongbuk production area and harvest time using UPLC-QTOF/MS and multivariate statistical analysis techniques. Journal of Applied Biological Chemistry. 67:113-122.
[https://doi.org/10.3839/jabc.2024.016]

- Song IG, An BR, Seo BI and Park SJ. (2009). Molecular marker to identify and origin of Cnidii Rhizoma from Korea and China. Korean Journal of Herbology. 24:1-8.
- Song JH, Yang S, Kim HB and Choi G. (2021). A comparative study about the origins of Apiaceae(Umbelliferae) taxa in the Pharmacopoeias of five Northeast-Asian countries based on the taxonomic concepts. Korean Journal of Herbology. 36:25-37.
- Suh Y, Kim YS, Lee C, Park J, Ko HJ, Lee SC, Jeong J and Choi HY. (2016). Taxonomic identity of leaf fragments found in the annals of the Joseon dynasty and botanical origin of a herbal medicine ‘Cheongung’. Korea Journal of Pharmacognosy. 47:128-136.
-
Um JN, Min JW, Joo KS and Kang HC. (2017). Antioxidant, anti-wrinkle activity and whitening effect of fermented mixture extracts of Angelica gigas, Paeonia Lactiflora, Rehmannia chinensis and Cnidium officinale. Korean Journal of Medicinal Crop Science. 25:152-159.
[https://doi.org/10.7783/KJMCS.2017.25.3.152]

-
Yamaki S, Ohyangi H, Yamasaki M, Eiguchi M, Miyabayashi T, Kubo T, Kurata N and Nonomura K. (2013). Development of INDEL markers to discriminate all genome types rapidly in the genus Oryza. Breeding Science. 63:246-254.
[https://doi.org/10.1270/jsbbs.63.246]

-
Yoon JB, Kwon DH and Jung CR. (2023). Analysis of wing forms and dominant of Thrips nigropilosus Uzel(Thysanoptera: Thripidae) inflicting to Cnidium officinale Makino. Korean Journal of Medicinal Crop Science. 31:316-323.
[https://doi.org/10.7783/KJMCS.2023.31.5.316]
